Mechanical Genomic Studies Reveal the Role of d-Alanine Metabolism in Pseudomonas aeruginosa Cell Stiffness
- PMID: 30206169
- PMCID: PMC6134093
- DOI: 10.1128/mBio.01340-18
Mechanical Genomic Studies Reveal the Role of d-Alanine Metabolism in Pseudomonas aeruginosa Cell Stiffness
Abstract
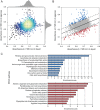
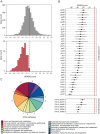
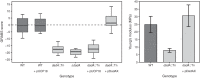
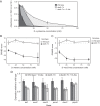
The stiffness of bacteria prevents cells from bursting due to the large osmotic pressure across the cell wall. Many successful antibiotic chemotherapies target elements that alter mechanical properties of bacteria, and yet a global view of the biochemistry underlying the regulation of bacterial cell stiffness is still emerging. This connection is particularly interesting in opportunistic human pathogens such as Pseudomonas aeruginosa that have a large (80%) proportion of genes of unknown function and low susceptibility to different families of antibiotics, including beta-lactams, aminoglycosides, and quinolones. We used a high-throughput technique to study a library of 5,790 loss-of-function mutants covering ~80% of the nonessential genes and correlated P. aeruginosa individual genes with cell stiffness. We identified 42 genes coding for proteins with diverse functions that, when deleted individually, decreased cell stiffness by >20%. This approach enabled us to construct a "mechanical genome" for P. aeruginosa d-Alanine dehydrogenase (DadA) is an enzyme that converts d-Ala to pyruvate that was included among the hits; when DadA was deleted, cell stiffness decreased by 18% (using multiple assays to measure mechanics). An increase in the concentration of d-Ala in cells downregulated the expression of genes in peptidoglycan (PG) biosynthesis, including the peptidoglycan-cross-linking transpeptidase genes ponA and dacC Consistent with this observation, ultraperformance liquid chromatography-mass spectrometry analysis of murein from P. aeruginosa cells revealed that dadA deletion mutants contained PG with reduced cross-linking and altered composition compared to wild-type cells.IMPORTANCE The mechanical properties of bacteria are important for protecting cells against physical stress. The cell wall is the best-characterized cellular element contributing to bacterial cell mechanics; however, the biochemistry underlying its regulation and assembly is still not completely understood. Using a unique high-throughput biophysical assay, we identified genes coding proteins that modulate cell stiffness in the opportunistic human pathogen Pseudomonas aeruginosa This approach enabled us to discover proteins with roles in a diverse range of biochemical pathways that influence the stiffness of P. aeruginosa cells. We demonstrate that d-Ala-a component of the peptidoglycan-is tightly regulated in cells and that its accumulation reduces expression of machinery that cross-links this material and decreases cell stiffness. This research demonstrates that there is much to learn about mechanical regulation in bacteria, and these studies revealed new nonessential P. aeruginosa targets that may enhance antibacterial chemotherapies or lead to new approaches.
Keywords: DadA; Pseudomonas aeruginosa; cell stiffness; cell wall; mechanical genomics.
Copyright © 2018 Trivedi et al.
Figures

Comment in
-
Who's Your DadA? d-Alanine Levels Regulate Bacterial Stiffness.mBio. 2018 Oct 23;9(5):e02127-18. doi: 10.1128/mBio.02127-18. mBio. 2018. PMID: 30352940 Free PMC article.
Similar articles
-
Who's Your DadA? d-Alanine Levels Regulate Bacterial Stiffness.mBio. 2018 Oct 23;9(5):e02127-18. doi: 10.1128/mBio.02127-18. mBio. 2018. PMID: 30352940 Free PMC article.
-
Identification of MupP as a New Peptidoglycan Recycling Factor and Antibiotic Resistance Determinant in Pseudomonas aeruginosa.mBio. 2017 Mar 28;8(2):e00102-17. doi: 10.1128/mBio.00102-17. mBio. 2017. PMID: 28351916 Free PMC article.
-
Cell-wall recycling and synthesis in Escherichia coli and Pseudomonas aeruginosa - their role in the development of resistance.J Med Microbiol. 2018 Jan;67(1):1-21. doi: 10.1099/jmm.0.000636. Epub 2017 Nov 29. J Med Microbiol. 2018. PMID: 29185941 Review.
-
A Proteolytic Complex Targets Multiple Cell Wall Hydrolases in Pseudomonas aeruginosa.mBio. 2018 Jul 17;9(4):e00972-18. doi: 10.1128/mBio.00972-18. mBio. 2018. PMID: 30018106 Free PMC article.
-
Bacterial Cell Mechanics.Biochemistry. 2017 Jul 25;56(29):3710-3724. doi: 10.1021/acs.biochem.7b00346. Epub 2017 Jul 11. Biochemistry. 2017. PMID: 28666084 Free PMC article. Review.
Cited by
-
Strategies and applications of antibacterial surface-modified biomaterials.Bioact Mater. 2025 Jul 9;53:114-140. doi: 10.1016/j.bioactmat.2025.07.009. eCollection 2025 Nov. Bioact Mater. 2025. PMID: 40688018 Free PMC article. Review.
-
Acinetobacter baumannii OmpA-like porins: functional characterization of bacterial physiology, antibiotic-resistance, and virulence.Commun Biol. 2024 Aug 6;7(1):948. doi: 10.1038/s42003-024-06645-0. Commun Biol. 2024. PMID: 39107399 Free PMC article.
-
Cell biomechanics and mechanobiology in bacteria: Challenges and opportunities.APL Bioeng. 2020 Apr 1;4(2):021501. doi: 10.1063/1.5135585. eCollection 2020 Jun. APL Bioeng. 2020. PMID: 32266323 Free PMC article. Review.
-
Bacterial Swarming Reduces Proteus mirabilis and Vibrio parahaemolyticus Cell Stiffness and Increases β-Lactam Susceptibility.mBio. 2019 Oct 8;10(5):e00210-19. doi: 10.1128/mBio.00210-19. mBio. 2019. PMID: 31594808 Free PMC article.
-
Variation of Burkholderia cenocepacia cell wall morphology and mechanical properties during cystic fibrosis lung infection, assessed by atomic force microscopy.Sci Rep. 2019 Nov 6;9(1):16118. doi: 10.1038/s41598-019-52604-9. Sci Rep. 2019. PMID: 31695169 Free PMC article.
References
-
- Stock JB, Rauch B, Roseman S. 1977. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem 252:7850–7861. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

