Geometric partitioning of cohesin and condensin is a consequence of chromatin loops
- PMID: 30207827
- PMCID: PMC6249845
- DOI: 10.1091/mbc.E18-02-0131
Geometric partitioning of cohesin and condensin is a consequence of chromatin loops
Abstract
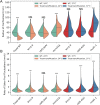
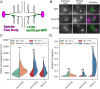
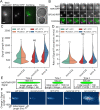
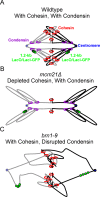
SMC (structural maintenance of chromosomes) complexes condensin and cohesin are crucial for proper chromosome organization. Condensin has been reported to be a mechanochemical motor capable of forming chromatin loops, while cohesin passively diffuses along chromatin to tether sister chromatids. In budding yeast, the pericentric region is enriched in both condensin and cohesin. As in higher-eukaryotic chromosomes, condensin is localized to the axial chromatin of the pericentric region, while cohesin is enriched in the radial chromatin. Thus, the pericentric region serves as an ideal model for deducing the role of SMC complexes in chromosome organization. We find condensin-mediated chromatin loops establish a robust chromatin organization, while cohesin limits the area that chromatin loops can explore. Upon biorientation, extensional force from the mitotic spindle aggregates condensin-bound chromatin from its equilibrium position to the axial core of pericentric chromatin, resulting in amplified axial tension. The axial localization of condensin depends on condensin's ability to bind to chromatin to form loops, while the radial localization of cohesin depends on cohesin's ability to diffuse along chromatin. The different chromatin-tethering modalities of condensin and cohesin result in their geometric partitioning in the presence of an extensional force on chromatin.
Figures




References
-
- Cohen J. (1988). Statistical Power Analysis for the Behavioral Sciences, Hillsdale, NJ: L. Erlbaum Associates.
-
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU. (2004). Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature , 93–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

