Layers of regulation of cell-cycle gene expression in the budding yeast Saccharomyces cerevisiae
- PMID: 30207828
- PMCID: PMC6249835
- DOI: 10.1091/mbc.E18-04-0255
Layers of regulation of cell-cycle gene expression in the budding yeast Saccharomyces cerevisiae
Abstract
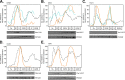
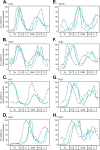
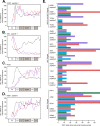
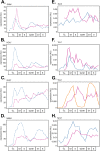
In the budding yeast Saccharomyces cerevisiae, transcription factors (TFs) regulate the periodic expression of many genes during the cell cycle, including gene products required for progression through cell-cycle events. Experimental evidence coupled with quantitative models suggests that a network of interconnected TFs is capable of regulating periodic genes over the cell cycle. Importantly, these dynamical models were built on transcriptomics data and assumed that TF protein levels and activity are directly correlated with mRNA abundance. To ask whether TF transcripts match protein expression levels as cells progress through the cell cycle, we applied a multiplexed targeted mass spectrometry approach (parallel reaction monitoring) to synchronized populations of cells. We found that protein expression of many TFs and cell-cycle regulators closely followed their respective mRNA transcript dynamics in cycling wild-type cells. Discordant mRNA/protein expression dynamics was also observed for a subset of cell-cycle TFs and for proteins targeted for degradation by E3 ubiquitin ligase complexes such as SCF (Skp1/Cul1/F-box) and APC/C (anaphase-promoting complex/cyclosome). We further profiled mutant cells lacking B-type cyclin/CDK activity ( clb1-6) where oscillations in ubiquitin ligase activity, cyclin/CDKs, and cell-cycle progression are halted. We found that a number of proteins were no longer periodically degraded in clb1-6 mutants compared with wild type, highlighting the importance of posttranscriptional regulation. Finally, the TF complexes responsible for activating G1/S transcription (SBF and MBF) were more constitutively expressed at the protein level than at periodic mRNA expression levels in both wild-type and mutant cells. This comprehensive investigation of cell-cycle regulators reveals that multiple layers of regulation (transcription, protein stability, and proteasome targeting) affect protein expression dynamics during the cell cycle.
Figures

Similar articles
-
Sirtuin 5 Is Regulated by the SCFCyclin F Ubiquitin Ligase and Is Involved in Cell Cycle Control.Mol Cell Biol. 2021 Jan 25;41(2):e00269-20. doi: 10.1128/MCB.00269-20. Print 2021 Jan 25. Mol Cell Biol. 2021. PMID: 33168699 Free PMC article.
-
Cdc20 protein contains a destruction-box but, unlike Clb2, its proteolysisis not acutely dependent on the activity of anaphase-promoting complex.Eur J Biochem. 2000 Jan;267(2):434-49. doi: 10.1046/j.1432-1327.2000.01014.x. Eur J Biochem. 2000. PMID: 10632713
-
Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression.Biochem J. 2007 Aug 1;405(3):569-81. doi: 10.1042/BJ20061812. Biochem J. 2007. PMID: 17461777 Free PMC article.
-
Ubiquitin-dependent proteolysis and cell cycle control in yeast.Prog Cell Cycle Res. 1996;2:115-27. doi: 10.1007/978-1-4615-5873-6_12. Prog Cell Cycle Res. 1996. PMID: 9552389 Review.
-
Structure, function and mechanism of the anaphase promoting complex (APC/C).Q Rev Biophys. 2011 May;44(2):153-90. doi: 10.1017/S0033583510000259. Epub 2010 Nov 22. Q Rev Biophys. 2011. PMID: 21092369 Review.
Cited by
-
A phosphatase-centric mechanism drives stress signaling response.EMBO Rep. 2021 Nov 4;22(11):e52476. doi: 10.15252/embr.202152476. Epub 2021 Sep 24. EMBO Rep. 2021. PMID: 34558777 Free PMC article.
-
Proteome-scale movements and compartment connectivity during the eukaryotic cell cycle.Cell. 2024 Mar 14;187(6):1490-1507.e21. doi: 10.1016/j.cell.2024.02.014. Epub 2024 Mar 6. Cell. 2024. PMID: 38452761 Free PMC article.
-
Control of meiotic entry by dual inhibition of a key mitotic transcription factor.Elife. 2024 Feb 27;12:RP90425. doi: 10.7554/eLife.90425. Elife. 2024. PMID: 38411169 Free PMC article.
-
Transcriptomic analysis of formic acid stress response in Saccharomyces cerevisiae.World J Microbiol Biotechnol. 2022 Jan 6;38(2):34. doi: 10.1007/s11274-021-03222-z. World J Microbiol Biotechnol. 2022. PMID: 34989900
-
Swi4-dependent SWI4 transcription couples cell size to cell cycle commitment.iScience. 2025 Feb 13;28(3):112027. doi: 10.1016/j.isci.2025.112027. eCollection 2025 Mar 21. iScience. 2025. PMID: 40124484 Free PMC article.
References
-
- Amon A, Tyers M, Futcher B, Nasmyth K. (1993). Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell , 993–1007. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

