A balance of deubiquitinating enzymes controls cell cycle entry
- PMID: 30207830
- PMCID: PMC6249862
- DOI: 10.1091/mbc.E18-07-0425
A balance of deubiquitinating enzymes controls cell cycle entry
Abstract
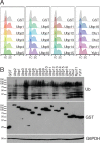
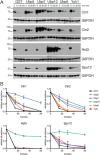
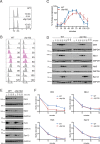
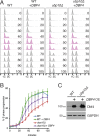
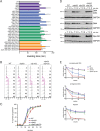
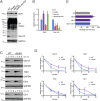
Protein degradation during the cell cycle is controlled by the opposing activities of ubiquitin ligases and deubiquitinating enzymes (DUBs). Although the functions of ubiquitin ligases in the cell cycle have been studied extensively, the roles of DUBs in this process are less well understood. Here, we used an overexpression screen to examine the specificities of each of the 21 DUBs in budding yeast for 37 cell cycle-regulated proteins. We find that DUBs up-regulate specific subsets of proteins, with five DUBs regulating the greatest number of targets. Overexpression of Ubp10 had the largest effect, stabilizing 15 targets and delaying cells in mitosis. Importantly, UBP10 deletion decreased the stability of the cell cycle regulator Dbf4, delayed the G1/S transition, and slowed proliferation. Remarkably, deletion of UBP10 together with deletion of four additional DUBs restored proliferation to near-wild-type levels. Among this group, deletion of the proteasome-associated DUB Ubp6 alone reversed the G1/S delay and restored the stability of Ubp10 targets in ubp10Δ cells. Similarly, deletion of UBP14, another DUB that promotes proteasomal activity, rescued the proliferation defect in ubp10Δ cells. Our results suggest that DUBs function through a complex genetic network in which their activities are coordinated to facilitate accurate cell cycle progression.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

