Insights on a putative aminoacyl-tRNA-protein transferase of Leishmania major
- PMID: 30208112
- PMCID: PMC6135404
- DOI: 10.1371/journal.pone.0203369
Insights on a putative aminoacyl-tRNA-protein transferase of Leishmania major
Abstract
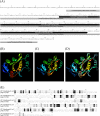
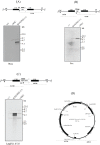
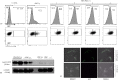
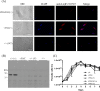
The N-end rule pathway leads to regulated proteolysis as an adaptive response to external stress and is ubiquitous from bacteria to mammals. In this study, we investigated a gene coding for a putative core enzyme of this post-translational regulatory pathway in Leishmania major, which may be crucial during cytodifferentiation and the environment adaptive responses of the parasite. Leucyl, phenylalanyl-tRNA protein transferase and arginyl-tRNA protein transferase are key components of this pathway in E. coli and eukaryotes, respectively. They catalyze the specific conjugation of leucine, phenylalanine or arginine to proteins containing exposed N-terminal amino acid residues, which are recognized by the machinery for the targeted proteolysis. Here, we characterized a conserved hypothetical protein coded by the LmjF.21.0725 gene in L. major. In silico analysis suggests that the LmjF.21.0725 protein is highly conserved among species of Leishmania and might belong to the Acyl CoA-N-acyltransferases (NAT) superfamily of proteins. Immunofluorescence cell imaging indicates that the cytosolic localization of the studied protein and the endogenous levels of the protein in promastigotes are barely detectable by western blotting assay. The knockout of the two alleles of LmjF.21.0725 by homologous recombination was only possible in the heterozygous transfectant expressing LmjF.21.0725 as a transgene from a plasmid. Moreover, the kinetics of loss of the plasmid in the absence of drug pressure suggests that maintenance of the gene is essential for promastigote survival. Here, evidence is provided that this putative aminoacyl tRNA-protein transferase is essential for parasite survival. The enzyme activity and corresponding post-translational regulatory pathway are yet to be investigated.
Conflict of interest statement
The authors have read the journal's policy and have the following conflict: Jena Bioscience GmbH provided support in the form of salary for R.B. and research materials. This does not alter our adherence to all the PLOS ONE policies on sharing data and materials.
Figures



Similar articles
-
The molecular basis for the post-translational addition of amino acids by L/F transferase in the N-end rule pathway.Curr Protein Pept Sci. 2015;16(2):163-80. Curr Protein Pept Sci. 2015. PMID: 25692952 Review.
-
Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog.EMBO J. 2006 Dec 13;25(24):5942-50. doi: 10.1038/sj.emboj.7601433. Epub 2006 Nov 16. EMBO J. 2006. PMID: 17110926 Free PMC article.
-
A reactive nucleophile proximal to vicinal thiols is an evolutionarily conserved feature in the mechanism of Arg aminoacyl-tRNA protein transferase.Arch Biochem Biophys. 1992 Nov 1;298(2):498-504. doi: 10.1016/0003-9861(92)90441-x. Arch Biochem Biophys. 1992. PMID: 1416979
-
C-terminal proteolysis of prenylated proteins in trypanosomatids and RNA interference of enzymes required for the post-translational processing pathway of farnesylated proteins.Mol Biochem Parasitol. 2007 Jun;153(2):115-24. doi: 10.1016/j.molbiopara.2007.02.009. Epub 2007 Mar 1. Mol Biochem Parasitol. 2007. PMID: 17397944
-
Post-translational N-terminal Arginylation of Protein Fragments: A Pivotal Portal to Proteolysis.Curr Protein Pept Sci. 2018;19(12):1214-1223. doi: 10.2174/1389203719666180809113122. Curr Protein Pept Sci. 2018. PMID: 30091410 Review.
Cited by
-
Structure and Mechanism of Aminoacyl-tRNA-Protein L/F- and R-transferases.J Mol Biol. 2025 Sep 1;437(17):169210. doi: 10.1016/j.jmb.2025.169210. Epub 2025 May 15. J Mol Biol. 2025. PMID: 40381981 Review.
References
-
- Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet (London, England). 2005;366 (9496):1561–77. - PubMed
-
- Glaser TA, Moody SF, Handman E, Bacic A, Spithill TW. An antigenically distinct lipophosphoglycan on amastigotes of Leishmania major. Mol Biochem Parasitol. 1991;45(2):337–44. - PubMed
-
- Garlapati S, Dahan E, Shapira M. Effect of acidic pH on heat shock gene expression in Leishmania. Mol Biochem Parasitol. 1999;100(1):95–101. - PubMed
-
- Quijada L, Soto M, Alonso C, Requena JM. Analysis of post-transcriptional regulation operating on transcription products of the tandemly linked Leishmania infantum hsp70 genes. J Biol Chem. 1997;272(7):4493–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

