The Zinc Linchpin Motif in the DNA Repair Glycosylase MUTYH: Identifying the Zn2+ Ligands and Roles in Damage Recognition and Repair
- PMID: 30208271
- PMCID: PMC6443246
- DOI: 10.1021/jacs.8b06923
The Zinc Linchpin Motif in the DNA Repair Glycosylase MUTYH: Identifying the Zn2+ Ligands and Roles in Damage Recognition and Repair
Abstract
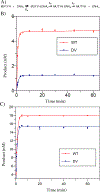
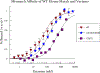
The DNA base excision repair (BER) glycosylase MUTYH prevents DNA mutations by catalyzing adenine (A) excision from inappropriately formed 8-oxoguanine (8-oxoG):A mismatches. The importance of this mutation suppression activity in tumor suppressor genes is underscored by the association of inherited variants of MUTYH with colorectal polyposis in a hereditary colorectal cancer syndrome known as MUTYH-associated polyposis, or MAP. Many of the MAP variants encompass amino acid changes that occur at positions surrounding the two-metal cofactor-binding sites of MUTYH. One of these cofactors, found in nearly all MUTYH orthologs, is a [4Fe-4S]2+ cluster coordinated by four Cys residues located in the N-terminal catalytic domain. We recently uncovered a second functionally relevant metal cofactor site present only in higher eukaryotic MUTYH orthologs: a Zn2+ ion coordinated by three Cys residues located within the extended interdomain connector (IDC) region of MUTYH that connects the N-terminal adenine excision and C-terminal 8-oxoG recognition domains. In this work, we identified a candidate for the fourth Zn2+ coordinating ligand using a combination of bioinformatics and computational modeling. In addition, using in vitro enzyme activity assays, fluorescence polarization DNA binding assays, circular dichroism spectroscopy, and cell-based rifampicin resistance assays, the functional impact of reduced Zn2+ chelation was evaluated. Taken together, these results illustrate the critical role that the "Zn2+ linchpin motif" plays in MUTYH repair activity by providing for proper engagement of the functional domains on the 8-oxoG:A mismatch required for base excision catalysis. The functional importance of the Zn2+ linchpin also suggests that adjacent MAP variants or exposure to environmental chemicals may compromise Zn2+ coordination, and ability of MUTYH to prevent disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

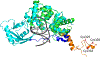
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

