Genetic Imbalances in Argentinean Patients with Congenital Conotruncal Heart Defects
- PMID: 30208644
- PMCID: PMC6162499
- DOI: 10.3390/genes9090454
Genetic Imbalances in Argentinean Patients with Congenital Conotruncal Heart Defects
Abstract
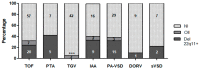
Congenital conotruncal heart defects (CCHD) are a subset of serious congenital heart defects (CHD) of the cardiac outflow tracts or great arteries. Its frequency is estimated in 1/1000 live births, accounting for approximately 10⁻30% of all CHD cases. Chromosomal abnormalities and copy number variants (CNVs) contribute to the disease risk in patients with syndromic and/or non-syndromic forms. Although largely studied in several populations, their frequencies are barely reported for Latin American countries. The aim of this study was to analyze chromosomal abnormalities, 22q11 deletions, and other genomic imbalances in a group of Argentinean patients with CCHD of unknown etiology. A cohort of 219 patients with isolated CCHD or associated with other major anomalies were referred from different provinces of Argentina. Cytogenetic studies, Multiplex-Ligation-Probe-Amplification (MLPA) and fluorescent in situ hybridization (FISH) analysis were performed. No cytogenetic abnormalities were found. 22q11 deletion was found in 23.5% of the patients from our cohort, 66% only had CHD with no other major anomalies. None of the patients with transposition of the great vessels (TGV) carried the 22q11 deletion. Other 4 clinically relevant CNVs were also observed: a distal low copy repeat (LCR)D-E 22q11 duplication, and 17p13.3, 4q35 and TBX1 deletions. In summary, 25.8% of CCHD patients presented imbalances associated with the disease.
Keywords: 22q11 deletion; conotruncal congenital heart defects; copy number variations.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures


References
-
- Pierpont M.E., Basson C.T., Benson D.W., Gelb B.D., Giglia T.M., Goldmuntz E., McGee G., Sable C.A., Srivastava D., Webb C.L. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the american heart association congenital cardiac defects committee, council on cardiovascular disease in the young: endorsed by the american academy of pediatrics. Circulation. 2007;115:3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. - DOI - PubMed
-
- Groisman B., Bidondo M.P., Duarte S., Piola A., Tardivo A., Liascovich R., Barbero P. Reporte Anual RENAC 2017. Análisis Epidemiológico Sobre las Anomalías Congénitas en Recién Nacidos, Registradas Durante 2016 en la República Argentina. [(accessed on 26 July 2018)]; Available online: http://www.msal.gob.ar/images/stories/bes/graficos/0000001111cnt-2018-02....
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

