Integrated human-virus metabolic stoichiometric modelling predicts host-based antiviral targets against Chikungunya, Dengue and Zika viruses
- PMID: 30209043
- PMCID: PMC6170780
- DOI: 10.1098/rsif.2018.0125
Integrated human-virus metabolic stoichiometric modelling predicts host-based antiviral targets against Chikungunya, Dengue and Zika viruses
Abstract
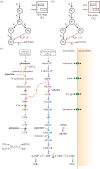
Current and reoccurring viral epidemic outbreaks such as those caused by the Zika virus illustrate the need for rapid development of antivirals. Such development would be facilitated by computational approaches that can provide experimentally testable predictions for possible antiviral strategies. To this end, we focus here on the fact that viruses are directly dependent on their host metabolism for reproduction. We develop a stoichiometric, genome-scale metabolic model that integrates human macrophage cell metabolism with the biochemical demands arising from virus production and use it to determine the virus impact on host metabolism and vice versa. While this approach applies to any host-virus pair, we first apply it to currently epidemic viruses Chikungunya, Dengue and Zika in this study. We find that each of these viruses causes specific alterations in the host metabolic flux towards fulfilling their biochemical demands as predicted by their genome and capsid structure. Subsequent analysis of this integrated model allows us to predict a set of host reactions, which, when constrained, inhibit virus production. We show that this prediction recovers known targets of existing antiviral drugs, specifically those targeting nucleotide production, while highlighting a set of hitherto unexplored reactions involving both amino acid and nucleotide metabolic pathways, with either broad or virus-specific antiviral potential. Thus, this computational approach allows rapid generation of experimentally testable hypotheses for novel antiviral targets within a host.
Keywords: antiviral targets; emerging viruses; flux balance analysis; host–virus interactions; metabolic modelling.
© 2018 The Authors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
A Novel Agonist of the TRIF Pathway Induces a Cellular State Refractory to Replication of Zika, Chikungunya, and Dengue Viruses.mBio. 2017 May 2;8(3):e00452-17. doi: 10.1128/mBio.00452-17. mBio. 2017. PMID: 28465426 Free PMC article.
-
Outbreak of Exanthematous Illness Associated with Zika, Chikungunya, and Dengue Viruses, Salvador, Brazil.Emerg Infect Dis. 2015 Dec;21(12):2274-6. doi: 10.3201/eid2112.151167. Emerg Infect Dis. 2015. PMID: 26584464 Free PMC article. No abstract available.
-
Carnosine exhibits significant antiviral activity against Dengue and Zika virus.J Pept Sci. 2019 Aug;25(8):e3196. doi: 10.1002/psc.3196. Epub 2019 Jul 9. J Pept Sci. 2019. PMID: 31290226
-
Dengue Virus Immunopathogenesis: Lessons Applicable to the Emergence of Zika Virus.J Mol Biol. 2016 Aug 28;428(17):3429-48. doi: 10.1016/j.jmb.2016.04.024. Epub 2016 Apr 27. J Mol Biol. 2016. PMID: 27130436 Review.
-
A review on structural genomics approach applied for drug discovery against three vector-borne viral diseases: Dengue, Chikungunya and Zika.Virus Genes. 2022 Jun;58(3):151-171. doi: 10.1007/s11262-022-01898-5. Epub 2022 Apr 8. Virus Genes. 2022. PMID: 35394596 Review.
Cited by
-
Genome-Scale Metabolic Model of Infection with SARS-CoV-2 Mutants Confirms Guanylate Kinase as Robust Potential Antiviral Target.Genes (Basel). 2021 May 24;12(6):796. doi: 10.3390/genes12060796. Genes (Basel). 2021. PMID: 34073716 Free PMC article.
-
Metabolic Model of the Phytophthora infestans-Tomato Interaction Reveals Metabolic Switches during Host Colonization.mBio. 2019 Jul 9;10(4):e00454-19. doi: 10.1128/mBio.00454-19. mBio. 2019. PMID: 31289172 Free PMC article.
-
Genome-scale model of Rothia mucilaginosa predicts gene essentialities and reveals metabolic capabilities.Microbiol Spectr. 2024 Jun 4;12(6):e0400623. doi: 10.1128/spectrum.04006-23. Epub 2024 Apr 23. Microbiol Spectr. 2024. PMID: 38652457 Free PMC article.
-
New workflow predicts drug targets against SARS-CoV-2 via metabolic changes in infected cells.PLoS Comput Biol. 2023 Mar 23;19(3):e1010903. doi: 10.1371/journal.pcbi.1010903. eCollection 2023 Mar. PLoS Comput Biol. 2023. PMID: 36952396 Free PMC article.
-
Integrating Systems and Synthetic Biology to Understand and Engineer Microbiomes.Annu Rev Biomed Eng. 2021 Jul 13;23:169-201. doi: 10.1146/annurev-bioeng-082120-022836. Epub 2021 Mar 29. Annu Rev Biomed Eng. 2021. PMID: 33781078 Free PMC article. Review.
References
-
- Merino-Ramos T, Vázquez-Calvo Á, Casas J, Sobrino F, Saiz J-C, Martín-Acebes MA. 2015. Modification of the host cell lipid metabolism induced by hypolipidemic drugs targeting the acetyl coenzyme A carboxylase impairs West Nile virus replication. Antimicrob. Agents Chemother. 60, 307–315. (10.1128/AAC.01578-15) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

