Reconstitution of immune cell interactions in free-standing membranes
- PMID: 30209137
- PMCID: PMC6398472
- DOI: 10.1242/jcs.219709
Reconstitution of immune cell interactions in free-standing membranes
Abstract
The spatiotemporal regulation of signalling proteins at the contacts formed between immune cells and their targets determines how and when immune responses begin and end. Therapeutic control of immune responses therefore relies on thorough elucidation of the molecular processes occurring at these interfaces. However, the detailed investigation of each component's contribution to the formation and regulation of the contact is hampered by the complexities of cell composition and architecture. Moreover, the transient nature of these interactions creates additional challenges, especially in the use of advanced imaging technology. One approach that circumvents these problems is to establish in vitro systems that faithfully mimic immune cell interactions, but allow complexity to be 'dialled-in' as needed. Here, we present an in vitro system that makes use of synthetic vesicles that mimic important aspects of immune cell surfaces. Using this system, we began to explore the spatial distribution of signalling molecules (receptors, kinases and phosphatases) and how this changes during the initiation of signalling. The GUV/cell system presented here is expected to be widely applicable.
Keywords: Giant unilamellar vesicles; Immune signalling; Immune synapse; In vitro reconstitution; Model membranes.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

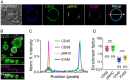


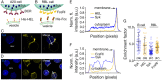
References
-
- Beningo K. A. and Wang Y. L. (2002). Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. J. Cell Sci. 115, 849-856. - PubMed
-
- Blouin C. M., Hamon Y., Gonnord P., Boularan C., Kagan J., Viaris de Lesegno C., Ruez R., Mailfert S., Bertaux N., Loew D. et al. (2016). Glycosylation-dependent IFN-gammaR partitioning in lipid and actin nanodomains is critical for JAK activation. Cell 166, 920-934. 10.1016/j.cell.2016.07.003 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

