Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression
- PMID: 30209170
- PMCID: PMC6232473
- DOI: 10.1128/JVI.01022-18
Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression
Abstract
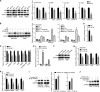
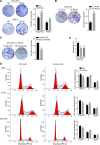
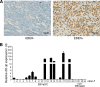
Epstein-Barr virus (EBV) is the first human virus found to encode many microRNAs. It is etiologically linked to nasopharyngeal carcinoma and EBV-associated gastric carcinoma. During the latent infection period, there are only a few EBV proteins expressed, whereas EBV microRNAs, such as the BamHI-A region rightward transcript (BART) microRNAs, are highly expressed. However, how these BART miRNAs precisely regulate the tumor growth in nasopharyngeal carcinoma and gastric carcinoma remains obscure. Here, we report that upregulation of EBV-miR-BART5-3p promotes the growth of nasopharyngeal carcinoma and gastric carcinoma cells. BART5-3p directly targets the tumor suppressor gene TP53 on its 3'-untranslated region (3'-UTR) and consequently downregulates CDKN1A, BAX, and FAS expression, leading to acceleration of the cell cycle progress and inhibition of cell apoptosis. BART5-3p contributes to the resistance to chemotherapeutic drugs and ionizing irradiation-induced p53 increase. Moreover, BART5-3p also facilitates degradation of p53 proteins. BART5-3p is the first EBV-microRNA to be identified as inhibiting p53 expression and function, which suggests a novel mechanism underlying the strategies employed by EBV to maintain latent infection and promote the development of EBV-associated carcinomas.IMPORTANCE EBV encodes 44 mature microRNAs, which have been proven to promote EBV-associated diseases by targeting host genes and self-viral genes. In EBV-associated carcinomas, the expression of viral protein is limited but the expression of BART microRNAs is extremely high, suggesting that they could be major factors in the contribution of EBV-associated tumorigenesis. p53 is a critical tumor suppressor. Unlike in most human solid tumors, TP53 mutations are rare in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues, suggesting a possibility that some EBV-encoded products suppress the functions of p53. This study provides the first evidence that a BART microRNA can suppress p53 expression by directly targeting its 3'-UTR. This study implies that EBV can use its BART microRNAs to modulate the expression of p53, thus maintaining its latency and contributing to tumorigenesis.
Keywords: Epstein-Barr virus; gastric cancer; miR-BART5-3p; nasopharyngeal carcinoma; p53.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
EBV-encoded miRNAs target ATM-mediated response in nasopharyngeal carcinoma.J Pathol. 2018 Apr;244(4):394-407. doi: 10.1002/path.5018. Epub 2018 Feb 16. J Pathol. 2018. PMID: 29230817 Free PMC article.
-
NF-κB Signaling Regulates Expression of Epstein-Barr Virus BART MicroRNAs and Long Noncoding RNAs in Nasopharyngeal Carcinoma.J Virol. 2016 Jun 24;90(14):6475-88. doi: 10.1128/JVI.00613-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27147748 Free PMC article.
-
Epstein-Barr virus-encoded miR-BART5-5p upregulates PD-L1 through PIAS3/pSTAT3 modulation, worsening clinical outcomes of PD-L1-positive gastric carcinomas.Gastric Cancer. 2020 Sep;23(5):780-795. doi: 10.1007/s10120-020-01059-3. Epub 2020 Mar 23. Gastric Cancer. 2020. PMID: 32206940
-
Epstein-Barr virus-encoded microRNAs as regulators in host immune responses.Int J Biol Sci. 2018 Apr 5;14(5):565-576. doi: 10.7150/ijbs.24562. eCollection 2018. Int J Biol Sci. 2018. PMID: 29805308 Free PMC article. Review.
-
Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases.Front Immunol. 2020 Mar 3;11:367. doi: 10.3389/fimmu.2020.00367. eCollection 2020. Front Immunol. 2020. PMID: 32194570 Free PMC article. Review.
Cited by
-
Implication of viral microRNAs in the genesis and diagnosis of Epstein-Barr virus-associated tumors.Oncol Lett. 2019 Oct;18(4):3433-3442. doi: 10.3892/ol.2019.10713. Epub 2019 Aug 5. Oncol Lett. 2019. PMID: 31516561 Free PMC article. Review.
-
Role of Exosomes and Their Potential as Biomarkers in Epstein-Barr Virus-Associated Gastric Cancer.Cancers (Basel). 2023 Jan 12;15(2):469. doi: 10.3390/cancers15020469. Cancers (Basel). 2023. PMID: 36672418 Free PMC article. Review.
-
Prognostic Epstein-Barr Virus (EBV) miRNA biomarkers for survival outcome in EBV-associated epithelial malignancies: Systematic review and meta-analysis.PLoS One. 2022 Apr 18;17(4):e0266893. doi: 10.1371/journal.pone.0266893. eCollection 2022. PLoS One. 2022. PMID: 35436288 Free PMC article.
-
Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs.Cancer Manag Res. 2019 Aug 27;11:8023-8031. doi: 10.2147/CMAR.S218967. eCollection 2019. Cancer Manag Res. 2019. PMID: 31695488 Free PMC article.
-
Epstein-Barr Virus miR-BART1-3p Regulates the miR-17-92 Cluster by Targeting E2F3.Int J Mol Sci. 2021 Oct 10;22(20):10936. doi: 10.3390/ijms222010936. Int J Mol Sci. 2021. PMID: 34681596 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

