Homologous recombination is an intrinsic defense against antiviral RNA interference
- PMID: 30209219
- PMCID: PMC6166822
- DOI: 10.1073/pnas.1810229115
Homologous recombination is an intrinsic defense against antiviral RNA interference
Abstract
RNA interference (RNAi) is the major antiviral defense mechanism of plants and invertebrates, rendering the capacity to evade it a defining factor in shaping the viral landscape. Here we sought to determine whether different virus replication strategies provided any inherent capacity to evade RNAi in the absence of an antagonist. Through the exploitation of host microRNAs, we recreated an RNAi-like environment in vertebrates and directly compared the capacity of positive- and negative-stranded RNA viruses to cope with this selective pressure. Applying this defense against four distinct viral families revealed that the capacity to undergo homologous recombination was the defining attribute that enabled evasion of this defense. Independent of gene expression strategy, positive-stranded RNA viruses that could undergo strand switching rapidly excised genomic material, while negative-stranded viruses were effectively targeted and cleared upon RNAi-based selection. These data suggest a dynamic relationship between host antiviral defenses and the biology of virus replication in shaping pathogen prevalence.
Keywords: RNAi; homologous recombination; miRNA; virus adaptation; virus polarity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
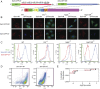
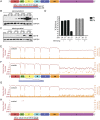

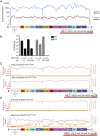
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

