Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer
- PMID: 30209313
- PMCID: PMC6135743
- DOI: 10.1038/s41467-018-06121-4
Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer
Abstract
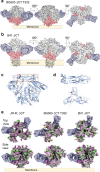
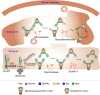
As the sole target of broadly neutralizing antibodies (bnAbs) to HIV, the envelope glycoprotein (Env) trimer is the focus of vaccination strategies designed to elicit protective bnAbs in humans. Because HIV Env is densely glycosylated with 75-90 N-glycans per trimer, most bnAbs use or accommodate them in their binding epitope, making the glycosylation of recombinant Env a key aspect of HIV vaccine design. Upon analysis of three HIV strains, we here find that site-specific glycosylation of Env from infectious virus closely matches Envs from corresponding recombinant membrane-bound trimers. However, viral Envs differ significantly from recombinant soluble, cleaved (SOSIP) Env trimers, strongly impacting antigenicity. These results provide a benchmark for virus Env glycosylation needed for the design of soluble Env trimers as part of an overall HIV vaccine strategy.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

