Thyroid hormone (T3) stimulates brown adipose tissue activation via mitochondrial biogenesis and MTOR-mediated mitophagy
- PMID: 30209975
- PMCID: PMC6287687
- DOI: 10.1080/15548627.2018.1511263
Thyroid hormone (T3) stimulates brown adipose tissue activation via mitochondrial biogenesis and MTOR-mediated mitophagy
Abstract
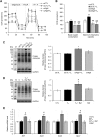
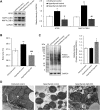
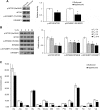
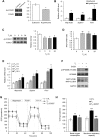
The thyroid hormone triiodothyronine (T3) activates thermogenesis by uncoupling electron transport from ATP synthesis in brown adipose tissue (BAT) mitochondria. Although T3 can induce thermogenesis by sympathetic innervation, little is known about its cell autonomous effects on BAT mitochondria. We thus examined effects of T3 on mitochondrial activity, autophagy, and metabolism in primary brown adipocytes and BAT and found that T3 increased fatty acid oxidation and mitochondrial respiration as well as autophagic flux, mitophagy, and mitochondrial biogenesis. Interestingly, there was no significant induction of intracellular reactive oxygen species (ROS) despite high mitochondrial respiration and UCP1 induction by T3. However, when cells were treated with Atg5 siRNA to block autophagy, induction of mitochondrial respiration by T3 decreased, and was accompanied by ROS accumulation, demonstrating a critical role for autophagic mitochondrial turnover. We next generated an Atg5 conditional knockout mouse model (Atg5 cKO) by injecting Ucp1 promoter-driven Cre-expressing adenovirus into Atg5Flox/Flox mice to examine effects of BAT-specific autophagy on thermogenesis in vivo. Hyperthyroid Atg5 cKO mice exhibited lower body temperature than hyperthyroid or euthyroid control mice. Metabolomic analysis showed that T3 increased short and long chain acylcarnitines in BAT, consistent with increased β-oxidation. T3 also decreased amino acid levels, and in conjunction with SIRT1 activation, decreased MTOR activity to stimulate autophagy. In summary, T3 has direct effects on mitochondrial autophagy, activity, and turnover in BAT that are essential for thermogenesis. Stimulation of BAT activity by thyroid hormone or its analogs may represent a potential therapeutic strategy for obesity and metabolic diseases. Abbreviations: ACACA: acetyl-Coenzyme A carboxylase alpha; AMPK: AMP-activated protein kinase; Acsl1: acyl-CoA synthetase long-chain family member 1; ATG5: autophagy related 5; ATG7: autophagy related 7; ATP: adenosine triphosphate; BAT: brown adipose tissue; cKO: conditional knockout; COX4I1: cytochrome c oxidase subunit 4I1; Cpt1b: carnitine palmitoyltransferase 1b, muscle; CQ: chloroquine; DAPI: 4',6-diamidino-2-phenylindole; DIO2: deiodinase, iodothyronine, type 2; DMEM: Dulbecco's modified Eagle's medium; EIF4EBP1: eukaryotic translation initiation factor 4E binding protein 1; Fabp4: fatty acid binding protein 4, adipocyte; FBS: fetal bovine serum; FCCP: carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone; FGF: fibroblast growth factor; FOXO1: forkhead box O1; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GFP: green fluorescent protein; Gpx1: glutathione peroxidase 1; Lipe: lipase, hormone sensitive; MAP1LC3B: microtubule-associated protein 1 light chain 3; mRNA: messenger RNA; MTORC1: mechanistic target of rapamycin kinase complex 1; NAD: nicotinamide adenine dinucleotide; Nrf1: nuclear respiratory factor 1; OCR: oxygen consumption rate; PBS: phosphate-buffered saline; PCR: polymerase chain reaction; PPARGC1A: peroxisome proliferative activated receptor, gamma, coactivator 1 alpha; Pnpla2: patatin-like phospholipase domain containing 2; Prdm16: PR domain containing 16; PRKA: protein kinase, AMP-activated; RPS6KB: ribosomal protein S6 kinase; RFP: red fluorescent protein; ROS: reactive oxygen species; SD: standard deviation; SEM: standard error of the mean; siRNA: small interfering RNA; SIRT1: sirtuin 1; Sod1: superoxide dismutase 1, soluble; Sod2: superoxide dismutase 2, mitochondrial; SQSTM1: sequestosome 1; T3: 3,5,3'-triiodothyronine; TFEB: transcription factor EB; TOMM20: translocase of outer mitochondrial membrane 20; UCP1: uncoupling protein 1 (mitochondrial, proton carrier); ULK1: unc-51 like kinase 1; VDAC1: voltage-dependent anion channel 1; WAT: white adipose tissue.
Keywords: Autophagy; brown adipose tissue; mitochondria; mitophagy; thermogenesis; thyroid hormone.
Figures
References
-
- Silva JE. The thermogenic effect of thyroid hormone and its clinical implications. Ann Intern Med. 2003. August 5;139(3):205–213. PubMed PMID: 12899588. - PubMed
-
- Bianco AC, Maia AL, da Silva WS, et al. Adaptive activation of thyroid hormone and energy expenditure. Biosci Rep. 2005. Jun-Aug;25(3–4):191–208. PubMed PMID: 16283553. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
