Complexity of Antibiotic Resistance in Commensal Escherichia coli Derived from Pigs from an Intensive-Production Farm
- PMID: 30210140
- PMCID: PMC6167118
- DOI: 10.1264/jsme2.ME17041
Complexity of Antibiotic Resistance in Commensal Escherichia coli Derived from Pigs from an Intensive-Production Farm
Abstract
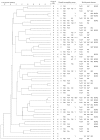
Antibiotics in animal husbandry are used to maintain welfare, but lead to the generation of resistant strains. We analyzed commensal multidrug-resistant Escherichia coli from pigs at the beginning and end of the production cycle in a farm with a farrow-to-finish system in order to investigate whether clonal spread or horizontal gene transfer constitutes the main factor responsible for the prevalence of resistance in this environment. Among 380 isolates, 56 multidrug-resistant E. coli with a similar resistant phenotype were selected for more detailed investigations including a genomic similarity analysis and the detection of mobile elements. Isolates carried blaTEM-1, aadA1, strA/B, tetA, tetB, tetC, dfrA1, dfrA5, dfrA7, dfrA12, sul1, sul2, sul3, and qnrS resistance genes, with the common co-occurrence of genes encoding the same resistance phenotype. A pulse-field gel electrophoresis analysis of the genomic similarity of multidrug-resistant E. coli showed ≤65% similarity of most of the tested strains and did not reveal a dominant clone responsible for the prevalence of resistance. Class 1 and 2 integrons and transposons 7 and 21 were detected among mobile elements; however, some were truncated. Plasmids were represented by 11 different incompatibility groups (K, FIB, I1, FIIA, FIC, FIA, Y, P, HI1, B/O, and T). Genetic resistance traits were unevenly spread in the clonal groups and suggested the major rearrangement of genetic material by horizontal gene transfer. The present results revealed that in commensal E. coli from pigs in a homogeneous farm environment, there was no dominant clone responsible for the spread of resistance and persistence in the population.
Keywords: antibiotic resistance; commensal Escherichia coli; food production animals.
Figures
References
-
- Ajiboye R.M., Solberg O.D., Lee B.M., Raphael E., DebRoy C., Riley L.W. Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin Infect Dis. 2009;49:365–371. - PubMed
-
- Bahl M.I., Hansen L.H., Sørensen S.J. Persistence mechanisms of conjugative plasmids. Methods Mol Biol (N Y, NY, U S) 2009;532:73–102. - PubMed
-
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. - PubMed
-
- Carattoli A. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol. 2011;301:654–658. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical