Post-transcriptional Processing of mRNA in Neurons: The Vestiges of the RNA World Drive Transcriptome Diversity
- PMID: 30210293
- PMCID: PMC6121099
- DOI: 10.3389/fnmol.2018.00304
Post-transcriptional Processing of mRNA in Neurons: The Vestiges of the RNA World Drive Transcriptome Diversity
Abstract
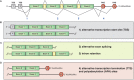
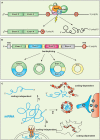
Neurons are morphologically complex cells that rely on the compartmentalization of protein expression to develop and maintain their extraordinary cytoarchitecture. This formidable task is achieved, at least in part, by targeting mRNA to subcellular compartments where they are rapidly translated. mRNA transcripts are the conveyor of genetic information from DNA to the translational machinery, however, they are also endowed with additional functions linked to both the coding sequence (open reading frame, or ORF) and the flanking 5' and 3' untranslated regions (UTRs), that may harbor coding-independent functions. In this review, we will highlight recent evidences supporting new coding-dependent and -independent functions of mRNA and discuss how nuclear and cytoplasmic post-transcriptional modifications of mRNA contribute to localization and translation in mammalian cells with specific emphasis on neurons. We also describe recently developed techniques that can be employed to study RNA dynamics at subcellular level in eukaryotic cells in developing and regenerating neurons.
Keywords: 3′UTR; RNA isoforms; RNA localization; RNA metabolism; RNA processing; neuron; translation.
Figures


References
-
- Andreassi C., Luisier R., Crerar H., Franke S., Luscombe M., Cuda G., et al. (2017). 3’UTR remodelling of axonal transcripts in sympathetic neurons. bioRxiv [Preprint] 10.1101/170100 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

