A General Protein O- Glycosylation Gene Cluster Encodes the Species-Specific Glycan of the Oral Pathogen Tannerella forsythia: O-Glycan Biosynthesis and Immunological Implications
- PMID: 30210478
- PMCID: PMC6120980
- DOI: 10.3389/fmicb.2018.02008
A General Protein O- Glycosylation Gene Cluster Encodes the Species-Specific Glycan of the Oral Pathogen Tannerella forsythia: O-Glycan Biosynthesis and Immunological Implications
Abstract
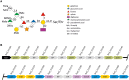
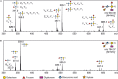
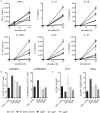
The cell surface of the oral pathogen Tannerella forsythia is heavily glycosylated with a unique, complex decasaccharide that is O-glycosidically linked to the bacterium's abundant surface (S-) layer, as well as other proteins. The S-layer glycoproteins are virulence factors of T. forsythia and there is evidence that protein O-glycosylation underpins the bacterium's pathogenicity. To elucidate the protein O-glycosylation pathway, genes suspected of encoding pathway components were first identified in the genome sequence of the ATCC 43037 type strain, revealing a 27-kb gene cluster that was shown to be polycistronic. Using a gene deletion approach targeted at predicted glycosyltransferases (Gtfs) and methyltransferases encoded in this gene cluster, in combination with mass spectrometry of the protein-released O-glycans, we show that the gene cluster encodes the species-specific part of the T. forsythia ATCC 43037 decasaccharide and that this is assembled step-wise on a pentasaccharide core. The core was previously proposed to be conserved within the Bacteroidetes phylum, to which T. forsythia is affiliated, and its biosynthesis is encoded elsewhere on the bacterial genome. Next, to assess the prevalence of protein O-glycosylation among Tannerella sp., the publicly available genome sequences of six T. forsythia strains were compared, revealing gene clusters of similar size and organization as found in the ATCC 43037 type strain. The corresponding region in the genome of a periodontal health-associated Tannerella isolate showed a different gene composition lacking most of the genes commonly found in the pathogenic strains. Finally, we investigated whether differential cell surface glycosylation impacts T. forsythia's overall immunogenicity. Release of proinflammatory cytokines by dendritic cells (DCs) upon stimulation with defined Gtf-deficient mutants of the type strain was measured and their T cell-priming potential post-stimulation was explored. This revealed that the O-glycan is pivotal to modulating DC effector functions, with the T. forsythia-specific glycan portion suppressing and the pentasaccharide core activating a Th17 response. We conclude that complex protein O-glycosylation is a hallmark of pathogenic T. forsythia strains and propose it as a valuable target for the design of novel antimicrobials against periodontitis.
Keywords: S-layer; carbohydrate-active enzymes; glycosyltransferase; immunogenicity; locus for glycosylation; methyltransferase; periodontitis.
Figures



Similar articles
-
Characterization of the O-Glycoproteome of Tannerella forsythia.mSphere. 2021 Oct 27;6(5):e0064921. doi: 10.1128/mSphere.00649-21. Epub 2021 Sep 15. mSphere. 2021. PMID: 34523981 Free PMC article.
-
Comparative genome characterization of the periodontal pathogen Tannerella forsythia.BMC Genomics. 2020 Feb 11;21(1):150. doi: 10.1186/s12864-020-6535-y. BMC Genomics. 2020. PMID: 32046654 Free PMC article.
-
Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia.J Biol Chem. 2011 Nov 4;286(44):38714-38724. doi: 10.1074/jbc.M111.284893. Epub 2011 Sep 12. J Biol Chem. 2011. PMID: 21911490 Free PMC article.
-
Nonulosonic acids contribute to the pathogenicity of the oral bacterium Tannerella forsythia.Interface Focus. 2019 Apr 6;9(2):20180064. doi: 10.1098/rsfs.2018.0064. Epub 2019 Feb 15. Interface Focus. 2019. PMID: 30842870 Free PMC article. Review.
-
Peptidoglycan Salvage Enables the Periodontal Pathogen Tannerella forsythia to Survive within the Oral Microbial Community.Microb Physiol. 2021;31(2):123-134. doi: 10.1159/000516751. Epub 2021 Jun 9. Microb Physiol. 2021. PMID: 34107471 Review.
Cited by
-
Glycosylation of oral bacteria in modulating adhesion and biofilm formation.J Oral Microbiol. 2025 Apr 8;17(1):2486650. doi: 10.1080/20002297.2025.2486650. eCollection 2025. J Oral Microbiol. 2025. PMID: 40213769 Free PMC article. Review.
-
Primary Structure of Glycans by NMR Spectroscopy.Chem Rev. 2023 Feb 8;123(3):1040-1102. doi: 10.1021/acs.chemrev.2c00580. Epub 2023 Jan 9. Chem Rev. 2023. PMID: 36622423 Free PMC article. Review.
-
Characterization of the O-Glycoproteome of Tannerella forsythia.mSphere. 2021 Oct 27;6(5):e0064921. doi: 10.1128/mSphere.00649-21. Epub 2021 Sep 15. mSphere. 2021. PMID: 34523981 Free PMC article.
-
Characterization of the O-Glycoproteome of Porphyromonas gingivalis.Microbiol Spectr. 2022 Feb 23;10(1):e0150221. doi: 10.1128/spectrum.01502-21. Epub 2022 Jan 5. Microbiol Spectr. 2022. PMID: 34985300 Free PMC article.
-
Activation of bone marrow-derived dendritic cells and CD4+ T cell differentiation by outer membrane vesicles of periodontal pathogens.J Oral Microbiol. 2022 Sep 14;14(1):2123550. doi: 10.1080/20002297.2022.2123550. eCollection 2022. J Oral Microbiol. 2022. PMID: 36312320 Free PMC article.
References
-
- Beall C. J., Campbell A. G., Dayeh D. M., Griffen A. L., Podar M., Leys E. J. (2014). Single cell genomics of uncultured, health-associated Tannerella BU063 (Oral Taxon 286) and comparison to the closely related pathogen Tannerella forsythia. PLoS One 9:e89398. 10.1371/journal.pone.0089398 - DOI - PMC - PubMed
-
- Bloch S., Zwicker S., Bostanci N., Sjöling A., Bostrom E. A., Belibasakis G. N., et al. (2018). Immune response profiling of primary monocytes and oral keratinocytes to different Tannerella forsythia strains and their cell surface mutants. Mol. Oral Microbiol. 33 155–167. 10.1111/omi.12208 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

