Impact of Chromosomal Architecture on the Function and Evolution of Bacterial Genomes
- PMID: 30210483
- PMCID: PMC6119826
- DOI: 10.3389/fmicb.2018.02019
Impact of Chromosomal Architecture on the Function and Evolution of Bacterial Genomes
Abstract
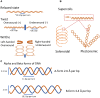
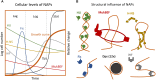
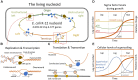
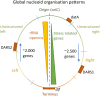
The bacterial nucleoid is highly condensed and forms compartment-like structures within the cell. Much attention has been devoted to investigating the dynamic topology and organization of the nucleoid. In contrast, the specific nucleoid organization, and the relationship between nucleoid structure and function is often neglected with regard to importance for adaption to changing environments and horizontal gene acquisition. In this review, we focus on the structure-function relationship in the bacterial nucleoid. We provide an overview of the fundamental properties that shape the chromosome as a structured yet dynamic macromolecule. These fundamental properties are then considered in the context of the living cell, with focus on how the informational flow affects the nucleoid structure, which in turn impacts on the genetic output. Subsequently, the dynamic living nucleoid will be discussed in the context of evolution. We will address how the acquisition of foreign DNA impacts nucleoid structure, and conversely, how nucleoid structure constrains the successful and sustainable chromosomal integration of novel DNA. Finally, we will discuss current challenges and directions of research in understanding the role of chromosomal architecture in bacterial survival and adaptation.
Keywords: bacterial nucleoid structure; chromosomal architecture; gene expression; genome evolution; nucleoid associated proteins.
Figures

References
-
- Bauer W. R. (1978). Structure and reactions.9114 of closed duplex dna. Annu. Rev. Biophys. Bioeng. 7 287–313. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

