Chimeric Proteins Containing MAP-1 and Functional Domains of C4b-Binding Protein Reveal Strong Complement Inhibitory Capacities
- PMID: 30210498
- PMCID: PMC6120983
- DOI: 10.3389/fimmu.2018.01945
Chimeric Proteins Containing MAP-1 and Functional Domains of C4b-Binding Protein Reveal Strong Complement Inhibitory Capacities
Abstract
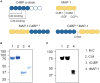
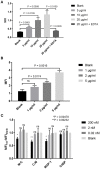
The complement system is a tightly regulated network of proteins involved in defense against pathogens, inflammatory processes, and coordination of the innate and adaptive immune responses. Dysregulation of the complement cascade is associated with many inflammatory disorders. Thus, inhibition of the complement system has emerged as an option for treatment of a range of different inflammatory diseases. MAP-1 is a pattern recognition molecule (PRM)-associated inhibitor of the lectin pathway of the complement system, whereas C4b-binding protein (C4BP) regulates both the classical and lectin pathways. In this study we generated chimeric proteins consisting of MAP-1 and the first five domains of human C4BP (C4BP1-5) in order to develop a targeted inhibitor acting at different levels of the complement cascade. Two different constructs were designed and expressed in CHO cells where MAP-1 was fused with C4BP1-5 in either the C- or N-terminus. The functionality of the chimeric proteins was assessed using different in vitro complement activation assays. Both chimeric proteins displayed the characteristic Ca2+-dependent dimerization and binding to PRMs of native MAP-1, as well as the co-factor activity of native C4BP. In ELISA-based complement activation assays they could effectively inhibit the lectin and classical pathways. Notably, MAP-1:C4BP1-5 was five times more effective than rMAP-1 and rC4BP1-5 applied at the same time, emphasizing the advantage of a single inhibitor containing both functional domains. The MAP-1/C4BP chimeras exert unique complement inhibitory properties and represent a novel therapeutic approach targeting both upstream and central complement activation.
Keywords: C4BP; MAP-1; chimeric protein; classical pathway; complement activation; complement inhibition; lectin pathway.
Figures






Similar articles
-
Combining MAP-1:CD35 or MAP-1:CD55 fusion proteins with pattern-recognition molecules as novel targeted modulators of the complement cascade.FASEB J. 2019 Nov;33(11):12723-12734. doi: 10.1096/fj.201901643R. Epub 2019 Aug 30. FASEB J. 2019. PMID: 31469600 Free PMC article.
-
Genetically engineered fusion of MAP-1 and factor H domains 1-5 generates a potent dual upstream inhibitor of both the lectin and alternative complement pathways.FASEB J. 2015 Dec;29(12):4945-55. doi: 10.1096/fj.15-277103. Epub 2015 Aug 10. FASEB J. 2015. PMID: 26260032
-
C4b Binding Protein Acts as an Innate Immune Effector Against Influenza A Virus.Front Immunol. 2021 Jan 8;11:585361. doi: 10.3389/fimmu.2020.585361. eCollection 2020. Front Immunol. 2021. PMID: 33488586 Free PMC article.
-
C4b-binding protein: The good, the bad and the deadly. Novel functions of an old friend.Immunol Lett. 2016 Jan;169:82-92. doi: 10.1016/j.imlet.2015.11.014. Epub 2015 Dec 2. Immunol Lett. 2016. PMID: 26658464 Review.
-
Diverse Functions of C4b-Binding Protein in Health and Disease.J Immunol. 2023 Nov 15;211(10):1443-1449. doi: 10.4049/jimmunol.2300333. J Immunol. 2023. PMID: 37931209 Free PMC article. Review.
Cited by
-
Combining MAP-1:CD35 or MAP-1:CD55 fusion proteins with pattern-recognition molecules as novel targeted modulators of the complement cascade.FASEB J. 2019 Nov;33(11):12723-12734. doi: 10.1096/fj.201901643R. Epub 2019 Aug 30. FASEB J. 2019. PMID: 31469600 Free PMC article.
-
PMMA-Based Continuous Hemofiltration Modulated Complement Activation and Renal Dysfunction in LPS-Induced Acute Kidney Injury.Front Immunol. 2021 Apr 1;12:605212. doi: 10.3389/fimmu.2021.605212. eCollection 2021. Front Immunol. 2021. PMID: 33868226 Free PMC article.
-
The alpha/B.1.1.7 SARS-CoV-2 variant exhibits significantly higher affinity for ACE-2 and requires lower inoculation doses to cause disease in K18-hACE2 mice.Elife. 2021 Nov 25;10:e70002. doi: 10.7554/eLife.70002. Elife. 2021. PMID: 34821555 Free PMC article.
-
Pathogenesis of Important Virulence Factors of Porphyromonas gingivalis via Toll-Like Receptors.Front Cell Infect Microbiol. 2019 Jul 18;9:262. doi: 10.3389/fcimb.2019.00262. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380305 Free PMC article. Review.
-
Functional Effects of Receptor-Binding Domain Mutations of SARS-CoV-2 B.1.351 and P.1 Variants.Front Immunol. 2021 Oct 7;12:757197. doi: 10.3389/fimmu.2021.757197. eCollection 2021. Front Immunol. 2021. PMID: 34691078 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

