The critical role of histone lysine demethylase KDM2B in cancer
- PMID: 30210666
- PMCID: PMC6129528
The critical role of histone lysine demethylase KDM2B in cancer
Abstract
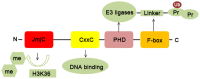
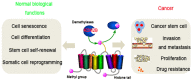
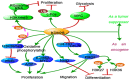
The discovery of histone demethylases has revealed the dynamic nature of the regulation of histone methylation. KDM2B is an important histone lysine demethylase that removes methyl from H3K36me2 and H3K4me3. It participates in many aspects of normal cellular processes such as cell senescence, cell differentiation and stem cell self-renewal. Recent studies also showed that KDM2B was overexpressed in various types of cancers. This review focuses primarily on the current knowledge of KDM2B and its function in cancer development.
Keywords: KDM2B; cancer; epigenetic regulation; histone demethylase.
Conflict of interest statement
None.
Figures
Similar articles
-
Histone demethylase KDM2B upregulates histone methyltransferase EZH2 expression and contributes to the progression of ovarian cancer in vitro and in vivo.Onco Targets Ther. 2017 Jun 26;10:3131-3144. doi: 10.2147/OTT.S134784. eCollection 2017. Onco Targets Ther. 2017. PMID: 28706445 Free PMC article.
-
Histone demethylase KDM2B promotes triple negative breast cancer proliferation by suppressing p15INK4B, p16INK4A, and p57KIP2 transcription.Acta Biochim Biophys Sin (Shanghai). 2018 Sep 1;50(9):897-904. doi: 10.1093/abbs/gmy084. Acta Biochim Biophys Sin (Shanghai). 2018. PMID: 30060056
-
Lysine demethylase 2B regulates angiogenesis via Jumonji C dependent suppression of angiogenic transcription factors.Biochem Biophys Res Commun. 2022 May 21;605:16-23. doi: 10.1016/j.bbrc.2022.03.054. Epub 2022 Mar 12. Biochem Biophys Res Commun. 2022. PMID: 35306360
-
A comprehensive review of lysine-specific demethylase 1 and its roles in cancer.Epigenomics. 2017 Aug;9(8):1123-1142. doi: 10.2217/epi-2017-0022. Epub 2017 Jul 12. Epigenomics. 2017. PMID: 28699367 Review.
-
Expanding the Role of the Histone Lysine-Specific Demethylase LSD1 in Cancer.Cancers (Basel). 2019 Mar 7;11(3):324. doi: 10.3390/cancers11030324. Cancers (Basel). 2019. PMID: 30866496 Free PMC article. Review.
Cited by
-
The Role of KDM2B and EZH2 in Regulating the Stemness in Colorectal Cancer Through the PI3K/AKT Pathway.Front Oncol. 2021 Mar 9;11:637298. doi: 10.3389/fonc.2021.637298. eCollection 2021. Front Oncol. 2021. PMID: 33791221 Free PMC article.
-
Systems-level identification of key transcription factors in immune cell specification.PLoS Comput Biol. 2022 Sep 26;18(9):e1010116. doi: 10.1371/journal.pcbi.1010116. eCollection 2022 Sep. PLoS Comput Biol. 2022. PMID: 36156073 Free PMC article.
-
The DNA/RNA helicase DHX9 orchestrates the KDM2B-mediated transcriptional regulation of YAP1 in Ewing sarcoma.Oncogene. 2024 Jan;43(4):225-234. doi: 10.1038/s41388-023-02894-1. Epub 2023 Nov 28. Oncogene. 2024. PMID: 38017132
-
Jumonji C Demethylases in Cellular Senescence.Genes (Basel). 2019 Jan 9;10(1):33. doi: 10.3390/genes10010033. Genes (Basel). 2019. PMID: 30634491 Free PMC article. Review.
-
Epigenetics of Cutaneous Sarcoma.Int J Mol Sci. 2021 Dec 31;23(1):422. doi: 10.3390/ijms23010422. Int J Mol Sci. 2021. PMID: 35008848 Free PMC article. Review.
References
-
- Elbjeirami WM. DNA methylation in the inflammatory response and relevance to chronic kidney disease. J Periodontol. 2008
-
- Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
