Genomic and expression profiling reveal molecular heterogeneity of disseminated tumor cells in bone marrow of early breast cancer
- PMID: 30211312
- PMCID: PMC6125436
- DOI: 10.1038/s41523-018-0083-5
Genomic and expression profiling reveal molecular heterogeneity of disseminated tumor cells in bone marrow of early breast cancer
Abstract
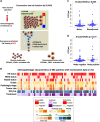
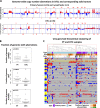
Detection of disseminated tumor cells (DTCs) in bone marrow is an established negative prognostic factor. We isolated small pools of (~20) EPCAM-positive DTCs from early breast cancer patients for genomic profiling. Genome-wide copy number profiles of DTC pools (n = 45) appeared less aberrant than the corresponding primary tumors (PT, n = 16). PIK3CA mutations were detected in 26% of DTC pools (n = 53), none of them were shared with matched PTs. Expression profiling of DTC pools (n = 30) confirmed the upregulation of EPCAM expression and certain oncogenes (e.g., MYC and CCNE1), as well as the absence of hematopoietic features. Two expression subtypes were observed: (1) luminal with dual epithelial-mesenchymal properties (high ESR1 and VIM/CAV1 expression), and (2) basal-like with proliferative/stem cell-like phenotype (low ESR1 and high MKI67/ALDH1A1 expression). We observed high discordance between ESR1 (40%) and ERRB2 (43%) expression in DTC pools vs. the clinical ER and HER2 status of the corresponding primary tumors, suggesting plasticity of biomarker status during dissemination to the bone marrow. Comparison of expression profiles of DTC pools with available data from circulating tumor cells (CTCs) of metastatic breast cancer patients revealed gene expression signatures in DTCs that were unique from those of CTCs. For example, ALDH1A1, CAV1, and VIM were upregulated in DTC pools relative to CTCs. Taken together, analysis of pooled DTCs revealed molecular heterogeneity, possible genetic divergence from corresponding primary tumor, and two distinct subpopulations. Validation in larger cohorts is needed to confirm the presence of these molecular subtypes and to evaluate their biological and clinical significance.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

