Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data
- PMID: 30211344
- PMCID: PMC6130812
- DOI: 10.1200/PO.17.00084
Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data
Abstract
Purpose: Microsatellite instability (MSI)/mismatch repair (MMR) status is increasingly important in the management of patients with cancer to predict response to immune checkpoint inhibitors. We determined MSI status from large-panel clinical targeted next-generation sequencing (NGS) data across various solid cancer types.
Methods: The MSI statuses of 12,288 advanced solid cancers consecutively sequenced with Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets clinical NGS assay were inferred by using MSIsensor, a program that reports the percentage of unstable microsatellites as a score. Cutoff score determination and sensitivity/specificity were based on MSI polymerase chain reaction (PCR) and MMR immunohistochemistry.
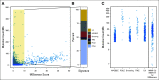
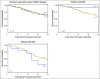
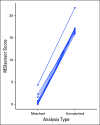
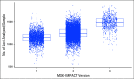
Results: By using an MSIsensor score ≥ 10 to define MSI high (MSI-H), 83 (8%) of 996 colorectal cancers (CRCs) and 42 (16%) of 260 uterine endometrioid cancers (UECs) were MSI-H. Validation against MSI PCR and/or MMR immunohistochemistry performed for 138 (24 MSI-H, 114 microsatellite stable [MSS]) CRCs, and 40 (15 MSI-H, 25 MSS) UECs showed a concordance of 99.4%. MSIsensor also identified 68 MSI-H/MMR-deficient (MMR-D) non-CRC/UECs. Of 9,591 non-CRC/UEC tumors with MSS MSIsensor status, 456 (4.8%) had slightly elevated scores(≥3 and <10) of which 96.6% with available material were confirmed to be MSS by MSI PCR. MSI-H was also detected and confirmed in three non-CRC/UECs with low exonic mutation burden (< 20). MSIsensor correctly scored all 15 polymerase ε ultra-mutated cancers as negative for MSI.
Conclusion: MSI status can be reliably inferred by MSIsensor from large-panel targeted NGS data. Concurrent MSI testing by NGS is resource efficient, is potentially more sensitive for MMR-D than MSI PCR, and allows identification of MSI-H across various cancers not typically screened, as highlighted by the finding that 35% (68 of 193) of all MSI-H tumors were non-CRC/ UEC.
Conflict of interest statement
Sumit Middha
No relationship to disclose
Liying Zhang
No relationship to disclose
Khedoudja Nafa
No relationship to disclose
Gowtham Jayakumaran
No relationship to disclose
Donna Wong
No relationship to disclose
Hyunjae R. Kim
No relationship to disclose
Justyna Sadowska
No relationship to disclose
Michael F. Berger
Deborah F. Delair
No relationship to disclose
Jinru Shia
No relationship to disclose
Zsofia Stadler
David S. Klimstra
Marc Ladanyi
Ahmet Zehir
No relationship to disclose
Jaclyn F. Hechtman
Figures




References
-
- Benson AB, III, Venook AP, Cederquist L, et al. : Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 15:370-398, 2017 - PubMed
-
- Hechtman JF, Middha S, Stadler ZK, et al: Universal screening for microsatellite instability in colorectal cancer in the clinical genomics era: new recommendations, methods, and considerations. Fam Cancer 10.1007/s10689-017-9993-x [epub ahead of print on April 12, 2017] - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

