Airway Microbiota Dynamics Uncover a Critical Window for Interplay of Pathogenic Bacteria and Allergy in Childhood Respiratory Disease
- PMID: 30212648
- PMCID: PMC6291254
- DOI: 10.1016/j.chom.2018.08.005
Airway Microbiota Dynamics Uncover a Critical Window for Interplay of Pathogenic Bacteria and Allergy in Childhood Respiratory Disease
Abstract
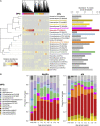
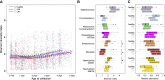
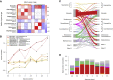
Repeated cycles of infection-associated lower airway inflammation drive the pathogenesis of persistent wheezing disease in children. In this study, the occurrence of acute respiratory tract illnesses (ARIs) and the nasopharyngeal microbiome (NPM) were characterized in 244 infants through their first five years of life. Through this analysis, we demonstrate that >80% of infectious events involve viral pathogens, but are accompanied by a shift in the NPM toward dominance by a small range of pathogenic bacterial genera. Unexpectedly, this change frequently precedes the detection of viral pathogens and acute symptoms. Colonization of illness-associated bacteria coupled with early allergic sensitization is associated with persistent wheeze in school-aged children, which is the hallmark of the asthma phenotype. In contrast, these bacterial genera are associated with "transient wheeze" that resolves after age 3 years in non-sensitized children. Thus, to complement early allergic sensitization, monitoring NPM composition may enable early detection and intervention in high-risk children.
Keywords: airway microbiota; allergic sensitization; asthma; lower respiratory infection.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
Breathe Soft, What Bugs through Early Windows Break?Cell Host Microbe. 2018 Sep 12;24(3):337-339. doi: 10.1016/j.chom.2018.08.012. Cell Host Microbe. 2018. PMID: 30212647
References
-
- Australian Commission on Safety and Quality in Health Care . ACSQHC; 2017. AURA 2017: Second Australian Report on Antimicrobial Use and Resistance in Human Health.
-
- Benjmini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300.
-
- Biesbroek G., Tsivtsivadze E., Sanders E.A., Montijn R., Veenhoven R.H., Keijser B.J., Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014;190:1283–1292. - PubMed
-
- Bisgaard H., Hermansen M.N., Buchvald F., Loland L., Halkjaer L.B., Bonnelykke K., Brasholt M., Heltberg A., Vissing N.H., Thorsen S.V. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007;357:1487–1495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

