Sizing up the cell cycle: systems and quantitative approaches in Chlamydomonas
- PMID: 30212737
- PMCID: PMC6269190
- DOI: 10.1016/j.pbi.2018.08.003
Sizing up the cell cycle: systems and quantitative approaches in Chlamydomonas
Abstract
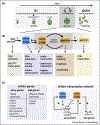
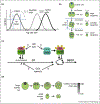
The unicellular green alga Chlamydomonas provides a simplified model for defining core cell cycle functions conserved in the green lineage and for understanding multiple fission, a common cell cycle variation found in many algae. Systems-level approaches including a recent groundbreaking screen for conditional lethal cell cycle mutants and genome-wide transcriptome analyses are revealing the complex relationships among cell cycle regulators and helping define roles for CDKA/CDK1 and CDKB, the latter of which is unique to the green lineage and plays a central role in mitotic regulation. Genetic screens and quantitative single-cell analyses have provided insight into cell-size control during multiple fission including the identification of a candidate `sizer' protein. Quantitative single-cell tracking and modeling are promising approaches for gaining additional insight into regulation of cellular and subcellular scaling during the Chlamydomonas cell cycle.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Cyclin-Dependent Kinase Regulation of Diurnal Transcription in Chlamydomonas.Plant Cell. 2015 Oct;27(10):2727-42. doi: 10.1105/tpc.15.00400. Epub 2015 Oct 16. Plant Cell. 2015. PMID: 26475866 Free PMC article.
-
Interregulation of CDKA/CDK1 and the Plant-Specific Cyclin-Dependent Kinase CDKB in Control of the Chlamydomonas Cell Cycle.Plant Cell. 2018 Feb;30(2):429-446. doi: 10.1105/tpc.17.00759. Epub 2018 Jan 24. Plant Cell. 2018. PMID: 29367304 Free PMC article.
-
A microbial avenue to cell cycle control in the plant superkingdom.Plant Cell. 2014 Oct;26(10):4019-38. doi: 10.1105/tpc.114.129312. Epub 2014 Oct 21. Plant Cell. 2014. PMID: 25336509 Free PMC article.
-
Chlamydomonas reinhardtii as a new model system for studying the molecular basis of the circadian clock.FEBS Lett. 2011 May 20;585(10):1495-502. doi: 10.1016/j.febslet.2011.02.025. Epub 2011 Feb 25. FEBS Lett. 2011. PMID: 21354416 Review.
-
New insights into the circadian clock in Chlamydomonas.Int Rev Cell Mol Biol. 2010;280:281-314. doi: 10.1016/S1937-6448(10)80006-1. Epub 2010 Mar 18. Int Rev Cell Mol Biol. 2010. PMID: 20797685 Review.
Cited by
-
The Papain-like Cysteine Protease HpXBCP3 from Haematococcus pluvialis Involved in the Regulation of Growth, Salt Stress Tolerance and Chlorophyll Synthesis in Microalgae.Int J Mol Sci. 2021 Oct 26;22(21):11539. doi: 10.3390/ijms222111539. Int J Mol Sci. 2021. PMID: 34768970 Free PMC article.
-
Starch granules in algal cells play an inherent role to shape the popular SSC signal in flow cytometry.BMC Res Notes. 2024 Oct 29;17(1):327. doi: 10.1186/s13104-024-06983-6. BMC Res Notes. 2024. PMID: 39472947 Free PMC article.
-
The biosynthesis of phospholipids is linked to the cell cycle in a model eukaryote.Biochim Biophys Acta Mol Cell Biol Lipids. 2021 Aug;1866(8):158965. doi: 10.1016/j.bbalip.2021.158965. Epub 2021 May 14. Biochim Biophys Acta Mol Cell Biol Lipids. 2021. PMID: 33992808 Free PMC article.
-
Hyperosmotic stress-induced microtubule disassembly in Chlamydomonas reinhardtii.BMC Plant Biol. 2022 Jan 22;22(1):46. doi: 10.1186/s12870-022-03439-6. BMC Plant Biol. 2022. PMID: 35065609 Free PMC article.
-
Bio-mining of Lanthanides from Red Mud by Green Microalgae.Molecules. 2019 Apr 6;24(7):1356. doi: 10.3390/molecules24071356. Molecules. 2019. PMID: 30959876 Free PMC article.
References
-
- Morgan DO: The Cell Cycle New Science Press Ltd; 2007.
-
- Harashima H, Dissmeyer N, Schnittger A: Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 2013, 23:345–356. - PubMed
-
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O: Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci 2012, 31:1-46.
-
- Harris E: Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol 2001, 52:363–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

