Synthetic negative feedback circuits using engineered small RNAs
- PMID: 30212900
- PMCID: PMC6182179
- DOI: 10.1093/nar/gky828
Synthetic negative feedback circuits using engineered small RNAs
Abstract
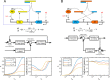
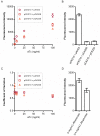
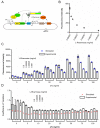
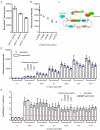
Negative feedback is known to enable biological and man-made systems to perform reliably in the face of uncertainties and disturbances. To date, synthetic biological feedback circuits have primarily relied upon protein-based, transcriptional regulation to control circuit output. Small RNAs (sRNAs) are non-coding RNA molecules that can inhibit translation of target messenger RNAs (mRNAs). In this work, we modelled, built and validated two synthetic negative feedback circuits that use rationally-designed sRNAs for the first time. The first circuit builds upon the well characterised tet-based autorepressor, incorporating an externally-inducible sRNA to tune the effective feedback strength. This allows more precise fine-tuning of the circuit output in contrast to the sigmoidal, steep input-output response of the autorepressor alone. In the second circuit, the output is a transcription factor that induces expression of an sRNA, which inhibits translation of the mRNA encoding the output, creating direct, closed-loop, negative feedback. Analysis of the noise profiles of both circuits showed that the use of sRNAs did not result in large increases in noise. Stochastic and deterministic modelling of both circuits agreed well with experimental data. Finally, simulations using fitted parameters allowed dynamic attributes of each circuit such as response time and disturbance rejection to be investigated.
Figures

Similar articles
-
A Modular Genetic System for High-Throughput Profiling and Engineering of Multi-Target Small RNAs.Methods Mol Biol. 2018;1737:373-391. doi: 10.1007/978-1-4939-7634-8_21. Methods Mol Biol. 2018. PMID: 29484604
-
The Phosphorolytic Exoribonucleases Polynucleotide Phosphorylase and RNase PH Stabilize sRNAs and Facilitate Regulation of Their mRNA Targets.J Bacteriol. 2016 Nov 18;198(24):3309-3317. doi: 10.1128/JB.00624-16. Print 2016 Dec 15. J Bacteriol. 2016. PMID: 27698082 Free PMC article.
-
Rational Design of High-Efficiency Synthetic Small Regulatory RNAs and Their Application in Robust Genetic Circuit Performance Through Tight Control of Leaky Gene Expression.ACS Synth Biol. 2024 Oct 18;13(10):3256-3267. doi: 10.1021/acssynbio.4c00323. Epub 2024 Sep 18. ACS Synth Biol. 2024. PMID: 39294875
-
Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria.Nat Rev Microbiol. 2016 Dec;14(12):775-784. doi: 10.1038/nrmicro.2016.129. Epub 2016 Sep 19. Nat Rev Microbiol. 2016. PMID: 27640758 Review.
-
Integration of Bacterial Small RNAs in Regulatory Networks.Annu Rev Biophys. 2017 May 22;46:131-148. doi: 10.1146/annurev-biophys-070816-034058. Annu Rev Biophys. 2017. PMID: 28532217 Review.
Cited by
-
Biomolecular mechanisms for signal differentiation.iScience. 2021 Nov 17;24(12):103462. doi: 10.1016/j.isci.2021.103462. eCollection 2021 Dec 17. iScience. 2021. PMID: 34927021 Free PMC article.
-
A cybergenetic framework for engineering intein-mediated integral feedback control systems.Nat Commun. 2023 Mar 11;14(1):1337. doi: 10.1038/s41467-023-36863-9. Nat Commun. 2023. PMID: 36906662 Free PMC article.
-
Double-edged role of resource competition in gene expression noise and control.Adv Genet (Hoboken). 2022 Mar;3(1):2100050. doi: 10.1002/ggn2.202100050. Epub 2022 Feb 8. Adv Genet (Hoboken). 2022. PMID: 35989723 Free PMC article.
-
Regulation of neutrophil myeloperoxidase inhibitor SPIN by the small RNA Teg49 in Staphylococcus aureus.Mol Microbiol. 2022 Jun;117(6):1447-1463. doi: 10.1111/mmi.14919. Epub 2022 May 31. Mol Microbiol. 2022. PMID: 35578788 Free PMC article.
-
Allosteric DNAzyme-based DNA logic circuit: operations and dynamic analysis.Nucleic Acids Res. 2019 Feb 20;47(3):1097-1109. doi: 10.1093/nar/gky1245. Nucleic Acids Res. 2019. PMID: 30541100 Free PMC article.
References
-
- Anderson J., Strelkowa N., Stan G.-B., Douglas T., Savulescu J., Barahona M., Papachristodoulou A.. Engineering and ethical perspectives in synthetic biology. Rigorous, robust and predictable designs, public engagement and a modern ethical framework are vital to the continued success of synthetic biology. EMBO Rep. 2012; 13:584–590. - PMC - PubMed
-
- Kwok R. Five hard truths for synthetic biology. Nature. 2010; 463:288–290. - PubMed
-
- Müller K.M., Arndt K.M.. Standardization in synthetic biology. Methods Mol. Biol. 2012; 813:23–43. - PubMed
-
- Canton B., Labno A., Endy D.. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008; 26:787–793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

