Snapshots of archaeal DNA replication and repair in living cells using super-resolution imaging
- PMID: 30212908
- PMCID: PMC6237752
- DOI: 10.1093/nar/gky829
Snapshots of archaeal DNA replication and repair in living cells using super-resolution imaging
Abstract
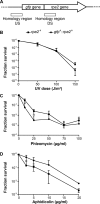
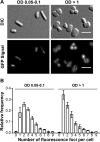
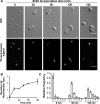
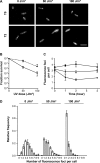
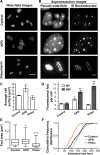
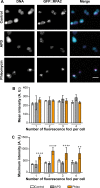
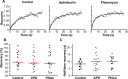
Using the haloarchaeon Haloferax volcanii as a model, we developed nascent DNA labeling and the functional GFP-labeled single-stranded binding protein RPA2 as novel tools to gain new insight into DNA replication and repair in live haloarchaeal cells. Our quantitative fluorescence microscopy data revealed that RPA2 forms distinct replication structures that dynamically responded to replication stress and DNA damaging agents. The number of the RPA2 foci per cell followed a probabilistic Poisson distribution, implying hitherto unnoticed stochastic cell-to-cell variation in haloarchaeal DNA replication and repair processes. The size range of haloarchaeal replication structures is very similar to those observed earlier in eukaryotic cells. The improved lateral resolution of 3D-SIM fluorescence microscopy allowed proposing that inhibition of DNA synthesis results in localized replication foci clustering and facilitated observation of RPA2 complexes brought about by chemical agents creating DNA double-strand breaks. Altogether our in vivo observations are compatible with earlier in vitro studies on archaeal single-stranded DNA binding proteins. Our work thus underlines the great potential of live cell imaging for unraveling the dynamic nature of transient molecular interactions that underpin fundamental molecular processes in the Third domain of life.
Figures
References
-
- Myllykallio H., Lopez P., Lopez-Garcia P., Heilig R., Saurin W., Zivanovic Y., Philippe H., Forterre P.. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000; 288:2212–2215. - PubMed
-
- Robinson N.P., Dionne I., Lundgren M., Marsh V.L., Bernander R., Bell S.D.. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004; 116:25–38. - PubMed
-
- Edgell D.R., Doolittle W.F.. Archaea and the origin(s) of DNA replication proteins. Cell. 1997; 89:995–998. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

