Characterization of the Microbial Population Inhabiting a Solar Saltern Pond of the Odiel Marshlands (SW Spain)
- PMID: 30213145
- PMCID: PMC6164061
- DOI: 10.3390/md16090332
Characterization of the Microbial Population Inhabiting a Solar Saltern Pond of the Odiel Marshlands (SW Spain)
Abstract
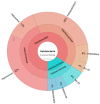
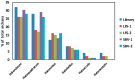
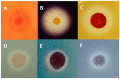
The solar salterns located in the Odiel marshlands, in southwest Spain, are an excellent example of a hypersaline environment inhabited by microbial populations specialized in thriving under conditions of high salinity, which remains poorly explored. Traditional culture-dependent taxonomic studies have usually under-estimated the biodiversity in saline environments due to the difficulties that many of these species have to grow at laboratory conditions. Here we compare two molecular methods to profile the microbial population present in the Odiel saltern hypersaline water ponds (33% salinity). On the one hand, the construction and characterization of two clone PCR amplified-16S rRNA libraries, and on the other, a high throughput 16S rRNA sequencing approach based on the Illumina MiSeq platform. The results reveal that both methods are comparable for the estimation of major genera, although massive sequencing provides more information about the less abundant ones. The obtained data indicate that Salinibacter ruber is the most abundant genus, followed by the archaea genera, Halorubrum and Haloquadratum. However, more than 100 additional species can be detected by Next Generation Sequencing (NGS). In addition, a preliminary study to test the biotechnological applications of this microbial population, based on its ability to produce and excrete haloenzymes, is shown.
Keywords: 16S rRNA metagenomics; Odiel marshlands; archaea; halo-extremophyles; haloenzymes.
Conflict of interest statement
“The authors declare no conflict of interest.” “The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript and in the decision to publish the results.”
Figures


Similar articles
-
Novel prokaryotic diversity in sediments of Tunisian multipond solar saltern.Res Microbiol. 2010 Sep;161(7):573-82. doi: 10.1016/j.resmic.2010.05.009. Epub 2010 Jun 15. Res Microbiol. 2010. PMID: 20558280
-
Prokaryotic diversity in Tuz Lake, a hypersaline environment in Inland Turkey.FEMS Microbiol Ecol. 2008 Sep;65(3):474-83. doi: 10.1111/j.1574-6941.2008.00510.x. Epub 2008 Jun 5. FEMS Microbiol Ecol. 2008. PMID: 18537839
-
Characterization of heterotrophic prokaryote subgroups in the Sfax coastal solar salterns by combining flow cytometry cell sorting and phylogenetic analysis.Extremophiles. 2011 May;15(3):347-58. doi: 10.1007/s00792-011-0364-5. Epub 2011 Mar 20. Extremophiles. 2011. PMID: 21424516 Free PMC article.
-
Microbial diversity of hypersaline environments: a metagenomic approach.Curr Opin Microbiol. 2015 Jun;25:80-7. doi: 10.1016/j.mib.2015.05.002. Epub 2015 Jun 8. Curr Opin Microbiol. 2015. PMID: 26056770 Review.
-
The Santa Pola saltern as a model for studying the microbiota of hypersaline environments.Extremophiles. 2014 Sep;18(5):811-24. doi: 10.1007/s00792-014-0681-6. Epub 2014 Aug 17. Extremophiles. 2014. PMID: 25129545 Review.
Cited by
-
Antioxidant, Antimicrobial, and Bioactive Potential of Two New Haloarchaeal Strains Isolated from Odiel Salterns (Southwest Spain).Biology (Basel). 2020 Sep 18;9(9):298. doi: 10.3390/biology9090298. Biology (Basel). 2020. PMID: 32962162 Free PMC article.
-
Up-Regulation of the Nrf2/HO-1 Antioxidant Pathway in Macrophages by an Extract from a New Halophilic Archaea Isolated in Odiel Saltworks.Antioxidants (Basel). 2023 May 11;12(5):1080. doi: 10.3390/antiox12051080. Antioxidants (Basel). 2023. PMID: 37237946 Free PMC article.
-
Transient Dynamics of Archaea and Bacteria in Sediments and Brine Across a Salinity Gradient in a Solar Saltern of Goa, India.Front Microbiol. 2020 Aug 13;11:1891. doi: 10.3389/fmicb.2020.01891. eCollection 2020. Front Microbiol. 2020. PMID: 33013726 Free PMC article.
-
Deep Subsurface Hypersaline Environment as a Source of Novel Species of Halophilic Sulfur-Oxidizing Bacteria.Microorganisms. 2022 May 9;10(5):995. doi: 10.3390/microorganisms10050995. Microorganisms. 2022. PMID: 35630438 Free PMC article.
-
Novel Halotolerant Bacteria from Saline Environments: Isolation and Biomolecule Production.BioTech (Basel). 2025 Jun 19;14(2):49. doi: 10.3390/biotech14020049. BioTech (Basel). 2025. PMID: 40558398 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

