Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids
- PMID: 30213835
- PMCID: PMC6365285
- DOI: 10.1158/2159-8290.CD-18-0474
Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids
Abstract
Based on genomic analysis, 50% of high-grade serous ovarian cancers (HGSC) are predicted to have DNA repair defects. Whether this substantial subset of HGSCs actually have functional repair defects remains unknown. Here, we devise a platform for functional profiling of DNA repair in short-term patient-derived HGSC organoids. We tested 33 organoid cultures derived from 22 patients with HGSC for defects in homologous recombination (HR) and replication fork protection. Regardless of DNA repair gene mutational status, a functional defect in HR in the organoids correlated with PARP inhibitor sensitivity. A functional defect in replication fork protection correlated with carboplatin and CHK1 and ATR inhibitor sensitivity. Our results indicate that a combination of genomic analysis and functional testing of organoids allows for the identification of targetable DNA damage repair defects. Larger numbers of patient-derived organoids must be analyzed to determine whether these assays can reproducibly predict patient response in the clinic.Significance: Patient-derived ovarian tumor organoids grow rapidly and match the tumors from which they are derived, both genetically and functionally. These organoids can be used for DNA repair profiling and therapeutic sensitivity testing and provide a rapid means of assessing targetable defects in the parent tumor, offering more suitable treatment options. Cancer Discov; 8(11); 1404-21. ©2018 AACR. This article is highlighted in the In This Issue feature, p. 1333.
©2018 American Association for Cancer Research.
Conflict of interest statement
For competing financial interests, two authors reported the following which is what Cancer Discovery reported for them: G.I. Shapiro reports receiving commercial research grants from Lilly, Sierra Oncology, and Merck/EMD Serono and is a consultant/advisory board member for Lilly, Sierra Oncology, Merck/EMD Serono, and Pfizer. A.D. D’Andrea reports receiving commercial research grants from Eli Lilly & Company, Sierra Oncology, and EMD Serono and is a consultant/advisory board member for Eli Lilly & Company, Sierra Oncology, and EMD Serono. No potential conflicts of interest were disclosed by the other authors.
Figures
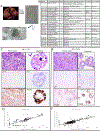





References
-
- Morphologic Ramalingam P., Immunophenotypic, and Molecular Features of Epithelial Ovarian Cancer. Oncology (Williston Park) 2016;30(2):166–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

