Membrane expression of thymidine kinase 1 and potential clinical relevance in lung, breast, and colorectal malignancies
- PMID: 30214377
- PMCID: PMC6131957
- DOI: 10.1186/s12935-018-0633-9
Membrane expression of thymidine kinase 1 and potential clinical relevance in lung, breast, and colorectal malignancies
Erratum in
-
Correction to: Membrane expression of thymidine kinase 1 and potential clinical relevance in lung, breast, and colorectal malignancies.Cancer Cell Int. 2019 Feb 11;19:29. doi: 10.1186/s12935-019-0749-6. eCollection 2019. Cancer Cell Int. 2019. PMID: 30792612 Free PMC article.
Abstract
Background: Lung, breast, and colorectal malignancies are the leading cause of cancer-related deaths in the world causing over 2.8 million cancer-related deaths yearly. Despite efforts to improve prevention methods, early detection, and treatments, survival rates for advanced stage lung, breast, and colon cancer remain low, indicating a critical need to identify cancer-specific biomarkers for early detection and treatment. Thymidine kinase 1 (TK1) is a nucleotide salvage pathway enzyme involved in cellular proliferation and considered an important tumor proliferation biomarker in the serum. In this study, we further characterized TK1's potential as a tumor biomarker and immunotherapeutic target and clinical relevance.
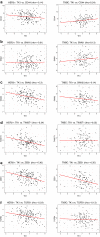
Methods: We assessed TK1 surface localization by flow cytometry and confocal microscopy in lung (NCI-H460, A549), breast (MDA-MB-231, MCF7), and colorectal (HT-29, SW620) cancer cell lines. We also isolated cell surface proteins from HT-29 cells and performed a western blot confirming the presence of TK1 on cell membrane protein fractions. To evaluate TK1's clinical relevance, we compared TK1 expression levels in normal and malignant tissue through flow cytometry and immunohistochemistry. We also analyzed RNA-Seq data from The Cancer Genome Atlas (TCGA) to assess differential expression of the TK1 gene in lung, breast, and colorectal cancer patients.
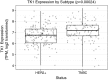
Results: We found significant expression of TK1 on the surface of NCI-H460, A549, MDA-MB-231, MCF7, and HT-29 cell lines and a strong association between TK1's localization with the membrane through confocal microscopy and Western blot. We found negligible TK1 surface expression in normal healthy tissue and significantly higher TK1 expression in malignant tissues. Patient data from TCGA revealed that the TK1 gene expression is upregulated in cancer patients compared to normal healthy patients.
Conclusions: Our results show that TK1 localizes on the surface of lung, breast, and colorectal cell lines and is upregulated in malignant tissues and patients compared to healthy tissues and patients. We conclude that TK1 is a potential clinical biomarker for the treatment of lung, breast, and colorectal cancer.
Keywords: Breast cancer; Colon cancer; Lung cancer; Membrane TK1; Surface expression; TK1; Thymidine kinase 1.
Figures








Similar articles
-
Novel monoclonal antibodies against thymidine kinase 1 and their potential use for the immunotargeting of lung, breast and colon cancer cells.Cancer Cell Int. 2020 Apr 16;20:127. doi: 10.1186/s12935-020-01198-8. eCollection 2020. Cancer Cell Int. 2020. PMID: 32317865 Free PMC article.
-
Thymidine kinase 1 through the ages: a comprehensive review.Cell Biosci. 2020 Nov 27;10(1):138. doi: 10.1186/s13578-020-00493-1. Cell Biosci. 2020. PMID: 33292474 Free PMC article. Review.
-
Non-small-cell lung cancer cell lines A549 and NCI-H460 express hypoxanthine guanine phosphoribosyltransferase on the plasma membrane.Onco Targets Ther. 2017 Mar 30;10:1921-1932. doi: 10.2147/OTT.S128416. eCollection 2017. Onco Targets Ther. 2017. PMID: 28408844 Free PMC article.
-
Breast and prostate cancer patients differ significantly in their serum Thymidine kinase 1 (TK1) specific activities compared with those hematological malignancies and blood donors: implications of using serum TK1 as a biomarker.BMC Cancer. 2015 Feb 18;15:66. doi: 10.1186/s12885-015-1073-8. BMC Cancer. 2015. PMID: 25881026 Free PMC article.
-
Comparative aspects of the proliferation marker thymidine kinase 1 in human and canine tumour diseases.Vet Comp Oncol. 2011 Mar;9(1):1-15. doi: 10.1111/j.1476-5829.2010.00238.x. Epub 2010 Aug 19. Vet Comp Oncol. 2011. PMID: 21303450 Review.
Cited by
-
Long noncoding RNA HOXC-AS3 indicates a poor prognosis and regulates tumorigenesis by binding to YBX1 in breast cancer.Am J Transl Res. 2020 Oct 15;12(10):6335-6350. eCollection 2020. Am J Transl Res. 2020. PMID: 33194034 Free PMC article.
-
Longitudinal single-cell profiling reveals molecular heterogeneity and tumor-immune evolution in refractory mantle cell lymphoma.Nat Commun. 2021 May 17;12(1):2877. doi: 10.1038/s41467-021-22872-z. Nat Commun. 2021. PMID: 34001881 Free PMC article.
-
Mechanisms of Resistance to CDK4/6 Inhibitors: Potential Implications and Biomarkers for Clinical Practice.Front Oncol. 2019 Jul 23;9:666. doi: 10.3389/fonc.2019.00666. eCollection 2019. Front Oncol. 2019. PMID: 31396487 Free PMC article. Review.
-
Computational Modelling of Tunicamycin C Interaction with Potential Protein Targets: Perspectives from Inverse Docking with Molecular Dynamic Simulation.Curr Issues Mol Biol. 2025 May 8;47(5):339. doi: 10.3390/cimb47050339. Curr Issues Mol Biol. 2025. PMID: 40699738 Free PMC article.
-
Novel monoclonal antibodies against thymidine kinase 1 and their potential use for the immunotargeting of lung, breast and colon cancer cells.Cancer Cell Int. 2020 Apr 16;20:127. doi: 10.1186/s12935-020-01198-8. eCollection 2020. Cancer Cell Int. 2020. PMID: 32317865 Free PMC article.
References
-
- Stewart BW, Wild CP. World cancer report 2014. World Health Organization. 2014. http://www.videnza.org/wp-content/uploads/World-Cancer-Report-2014.pdf. Accessed 2 Sept 2016.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

