Holding All the CARDs: How MALT1 Controls CARMA/CARD-Dependent Signaling
- PMID: 30214442
- PMCID: PMC6125328
- DOI: 10.3389/fimmu.2018.01927
Holding All the CARDs: How MALT1 Controls CARMA/CARD-Dependent Signaling
Abstract
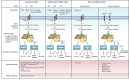
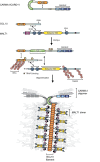
The scaffold proteins CARMA1-3 (encoded by the genes CARD11, -14 and -10) and CARD9 play major roles in signaling downstream of receptors with immunoreceptor tyrosine activation motifs (ITAMs), G-protein coupled receptors (GPCR) and receptor tyrosine kinases (RTK). These receptors trigger the formation of oligomeric CARMA/CARD-BCL10-MALT1 (CBM) complexes via kinases of the PKC family. The CBM in turn regulates gene expression by the activation of NF-κB and AP-1 transcription factors and controls transcript stability. The paracaspase MALT1 is the only CBM component having an enzymatic (proteolytic) activity and has therefore recently gained attention as a potential drug target. Here we review recent advances in the understanding of the molecular function of the protease MALT1 and summarize how MALT1 scaffold and protease function contribute to the transmission of CBM signals. Finally, we will highlight how dysregulation of MALT1 function can cause pathologies such as immunodeficiency, autoimmunity, psoriasis, and cancer.
Keywords: BCR; EGFR; GPCR; RNA stability; TCR; Treg; paracaspase; ubiquitin.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

