The autophagy receptor SQSTM1/p62 mediates anti-inflammatory actions of the selective NR3C1/glucocorticoid receptor modulator compound A (CpdA) in macrophages
- PMID: 30215534
- PMCID: PMC6984772
- DOI: 10.1080/15548627.2018.1495681
The autophagy receptor SQSTM1/p62 mediates anti-inflammatory actions of the selective NR3C1/glucocorticoid receptor modulator compound A (CpdA) in macrophages
Abstract
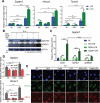
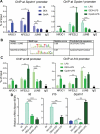
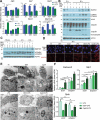
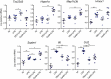
Glucocorticoids are widely used to treat inflammatory disorders; however, prolonged use of glucocorticoids results in side effects including osteoporosis, diabetes and obesity. Compound A (CpdA), identified as a selective NR3C1/glucocorticoid receptor (nuclear receptor subfamily 3, group C, member 1) modulator, exhibits an inflammation-suppressive effect, largely in the absence of detrimental side effects. To understand the mechanistic differences between the classic glucocorticoid dexamethasone (DEX) and CpdA, we looked for proteins oppositely regulated in bone marrow-derived macrophages using an unbiased proteomics approach. We found that the autophagy receptor SQSTM1 but not NR3C1 mediates the anti-inflammatory action of CpdA. CpdA drives SQSTM1 upregulation by recruiting the NFE2L2 transcription factor to its promoter. In contrast, the classic NR3C1 ligand dexamethasone recruits NR3C1 to the Sqstm1 promoter and other NFE2L2-controlled gene promoters, resulting in gene downregulation. Both DEX and CpdA induce autophagy, with marked different autophagy characteristics and morphology. Suppression of LPS-induced Il6 and Ccl2 genes by CpdA in macrophages is hampered upon Sqstm1 silencing, confirming that SQSTM1 is essential for the anti-inflammatory capacity of CpdA, at least in this cell type. Together, these results demonstrate how off-target mechanisms of selective NR3C1 ligands may contribute to a more efficient anti-inflammatory therapy.
Keywords: Autophagy; CpdA; NFE2L2/NRF2; SQSTM1/p62; autophagy receptors; glucocorticoids; inflammation.
Figures


References
-
- Chatzidionysiou K, Emamikia S, Nam J, et al. Efficacy of glucocorticoids, conventional and targeted synthetic disease-modifying antirheumatic drugs: A systematic literature review informing the 2016 update of the EULAR recommendations for the management of rheumatoid arthritis. Ann Rheum Dis. 2017;76:1102–1107. - PubMed
-
- Barnes PJ. Glucocorticosteroids. Handb Exp Pharmacol. 2016;237:93–115. - PubMed
-
- Ratman D, Vanden Berghe W, Dejager L, et al. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol Cell Endocrinol. 2013;380:41–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
