An L,L-diaminopimelate aminotransferase mutation leads to metabolic shifts and growth inhibition in Arabidopsis
- PMID: 30215754
- PMCID: PMC6255705
- DOI: 10.1093/jxb/ery325
An L,L-diaminopimelate aminotransferase mutation leads to metabolic shifts and growth inhibition in Arabidopsis
Abstract
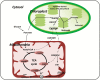
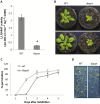
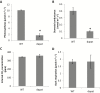
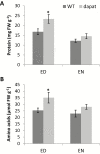
Lysine (Lys) connects the mitochondrial electron transport chain to amino acid catabolism and the tricarboxylic acid cycle. However, our understanding of how a deficiency in Lys biosynthesis impacts plant metabolism and growth remains limited. Here, we used a previously characterized Arabidopsis mutant (dapat) with reduced activity of the Lys biosynthesis enzyme L,L-diaminopimelate aminotransferase to investigate the physiological and metabolic impacts of impaired Lys biosynthesis. Despite displaying similar stomatal conductance and internal CO2 concentration, we observed reduced photosynthesis and growth in the dapat mutant. Surprisingly, whilst we did not find differences in dark respiration between genotypes, a lower storage and consumption of starch and sugars was observed in dapat plants. We found higher protein turnover but no differences in total amino acids during a diurnal cycle in dapat plants. Transcriptional and two-dimensional (isoelectric focalization/SDS-PAGE) proteome analyses revealed alterations in the abundance of several transcripts and proteins associated with photosynthesis and photorespiration coupled with a high glycine/serine ratio and increased levels of stress-responsive amino acids. Taken together, our findings demonstrate that biochemical alterations rather than stomatal limitations are responsible for the decreased photosynthesis and growth of the dapat mutant, which we hypothesize mimics stress conditions associated with impairments in the Lys biosynthesis pathway.
Figures




References
-
- Angelovici R, Fait A, Fernie AR, Galili G. 2011. A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytologist 189, 148–159. - PubMed
-
- Araújo WL, Nunes-Nesi A, Nikoloski Z, Sweetlove LJ, Fernie AR. 2012. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant, Cell & Environment 35, 1–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

