CRISPR-Cap: multiplexed double-stranded DNA enrichment based on the CRISPR system
- PMID: 30215766
- PMCID: PMC6326800
- DOI: 10.1093/nar/gky820
CRISPR-Cap: multiplexed double-stranded DNA enrichment based on the CRISPR system
Abstract
Existing methods to enrich target regions of genomic DNA based on PCR, hybridization capture, or molecular inversion probes have various drawbacks, including long experiment times and low throughput and/or enrichment quality. We developed CRISPR-Cap, a simple and scalable CRISPR-based method to enrich target regions of dsDNA, requiring only two short experimental procedures that can be completed within two hours. We used CRISPR-Cap to enrich 10 target genes 355.7-fold on average from Escherichia coli genomic DNA with a maximum on-target ratio of 81% and high enrichment uniformity. We also used CRISPR-Cap to measure gene copy numbers and detect rare alleles with frequencies as low as 1%. Finally, we enriched coding sequence regions of 20 genes from the human genome. We envision that CRISPR-Cap can be used as an alternative to other widely used target-enrichment methods, which will broaden the scope of CRISPR applications to the field of target enrichment field.
Figures
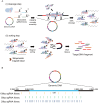
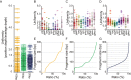
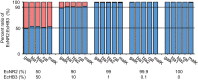
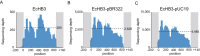

References
-
- Henegariu O., Heerema N.A., Dlouhy S.R., Vance G.H., Vogt P.H.. Multiplex PCR: Critical parameters and step-by-step protocol. BioTechniques. 1997; 23:504–511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

