Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016
- PMID: 30216671
- PMCID: PMC6304321
- DOI: 10.1111/irv.12610
Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016
Abstract
Background: Six amino acid positions (145, 155, 156, 158, 159, and 189, referred to as the antigenic motif; H3 numbering) in the globular head region of hemagglutinin (HA1 domain) play an important role in defining the antigenic phenotype of swine Clade IV (C-IV) H3N2 IAV, containing an H3 from a late 1990s human-to-swine introduction. We hypothesized that antigenicity of a swine C-IV H3 virus could be inferred based upon the antigenic motif if it matched a previously characterized antigen with the same motif. An increasing number of C-IV H3 genes encoding antigenic motifs that had not been previously characterized were observed in the U.S. pig population between 2012 and 2016.
Objectives: A broad panel of contemporary H3 viruses with uncharacterized antigenic motifs was selected across multiple clades within C-IV to assess the impact of HA1 genetic diversity on the antigenic phenotype.
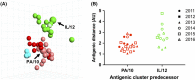
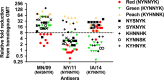
Methods: Hemagglutination inhibition (HI) assays were performed with isolates selected based on antigenic motif, tested against a panel of swine antisera, and visualized by antigenic cartography.
Results: A previously uncharacterized motif with low but sustained circulation in the swine population demonstrated a distinct phenotype from those previously characterized. Antigenic variation increased for viruses with similar antigenic motifs, likely due to amino acid substitutions outside the motif.
Conclusions: Although antigenic motifs were largely associated with antigenic distances, substantial diversity among co-circulating viruses poses a significant challenge for effective vaccine development. Continued surveillance and antigenic characterization of circulating strains is critical for improving vaccine efforts to control C-IV H3 IAV in U.S. swine.
Keywords: H3N2; antigenic cartography; antigenic evolution; influenza A virus; swine.
© 2018 The Authors. Influenza and Other Respiratory Viruses Published by John Wiley & Sons Ltd.
Figures



References
-
- World Health Organization . Consultations on the composition of influenza virus vaccines.
-
- Zoetis . FluSure XP. https://www.zoetisus.com/products/pork/flusure-xp/index.aspx. Accessed June 20, 2017.
-
- Center for Veterinary Biologics . Notice N. 17‐01. 2017.
-
- Center for Veterinary Biologics . Notice N. 17‐09. 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

