Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview
- PMID: 30217093
- PMCID: PMC6165440
- DOI: 10.3390/v10090497
Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview
Abstract
Influenza A viruses (IAVs) possess a great zoonotic potential as they are able to infect different avian and mammalian animal hosts, from which they can be transmitted to humans. This is based on the ability of IAV to gradually change their genome by mutation or even reassemble their genome segments during co-infection of the host cell with different IAV strains, resulting in a high genetic diversity. Variants of circulating or newly emerging IAVs continue to trigger global health threats annually for both humans and animals. Here, we provide an introduction on IAVs, highlighting the mechanisms of viral evolution, the host spectrum, and the animal/human interface. Pathogenicity determinants of IAVs in mammals, with special emphasis on newly emerging IAVs with pandemic potential, are discussed. Finally, an overview is provided on various approaches for the prevention of human IAV infections.
Keywords: Influenza A virus; evolution; pandemic; pathogenicity; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

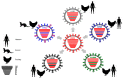
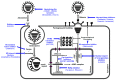
References
-
- Shaw M.L., Palese P. Orthomyxoviridae. 6th ed. Volume 1. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1151–1185.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

