A spontaneous mouse deletion in Mctp1 uncovers a long-range cis-regulatory region crucial for NR2F1 function during inner ear development
- PMID: 30217595
- PMCID: PMC6214362
- DOI: 10.1016/j.ydbio.2018.09.011
A spontaneous mouse deletion in Mctp1 uncovers a long-range cis-regulatory region crucial for NR2F1 function during inner ear development
Abstract
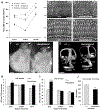
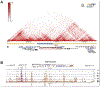
Hundreds of thousands of cis-regulatory DNA sequences are predicted in vertebrate genomes, but unlike genes themselves, few have been characterized at the functional level or even unambiguously paired with a target gene. Here we serendipitously identified and started investigating the first reported long-range regulatory region for the Nr2f1 (Coup-TFI) transcription factor gene. NR2F1 is temporally and spatially regulated during development and required for patterning and regionalization in the nervous system, including sensory hair cell organization in the auditory epithelium of the cochlea. Analyzing the deaf wanderer (dwnd) spontaneous mouse mutation, we traced back the cause of its associated circling behavior to a 53 kb deletion removing five exons and adjacent intronic regions of the poorly characterized Mctp1 gene. Interestingly, loss of Mctp1 function cannot account for the hearing loss, inner ear dysmorphology and sensory hair cell disorganization observed in dwnd mutants. Instead, we found that the Mctp1dwnd deletion affects the Nr2f1 gene located 1.4 Mb away, downregulating transcription and protein expression in the embryonic cochlea. Remarkably, the Mctp1dwnd allele failed to complement a targeted inactivation allele of Nr2f1, and transheterozygotes or Mctp1dwnd homozygotes exhibit the same morphological defects observed in inner ears of Nr2f1 mutants without sharing their early life lethality. Defects include improper separation of the utricle and saccule in the vestibule not described previously, which can explain the circling behavior that first brought the spontaneous mutation to attention. By contrast, mice homozygous for a targeted inactivation of Mctp1 have normal hearing and inner ear structures. We conclude that the 53 kb Mctp1dwnd deletion encompasses a long-range cis-regulatory region essential for proper Nr2f1 expression in the embryonic inner ear, providing a first opportunity to investigate Nr2f1 function in postnatal inner ears. This work adds to the short list of long-range regulatory regions characterized as essential to drive expression of key developmental control genes.
Keywords: Cis-regulation; Deafness; Inner ear development; Long-range enhancer; Mctp1; Nr2f1.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Deletion of a Long-Range Dlx5 Enhancer Disrupts Inner Ear Development in Mice.Genetics. 2018 Mar;208(3):1165-1179. doi: 10.1534/genetics.117.300447. Epub 2018 Jan 3. Genetics. 2018. PMID: 29301908 Free PMC article.
-
A coregulatory network of NR2F1 and microRNA-140.PLoS One. 2013 Dec 3;8(12):e83358. doi: 10.1371/journal.pone.0083358. eCollection 2013. PLoS One. 2013. PMID: 24349493 Free PMC article.
-
Multiple enhancers located in a 1-Mb region upstream of POU3F4 promote expression during inner ear development and may be required for hearing.Hum Genet. 2010 Oct;128(4):411-9. doi: 10.1007/s00439-010-0864-x. Epub 2010 Jul 29. Hum Genet. 2010. PMID: 20668882 Free PMC article.
-
MicroRNAs and epigenetic regulation in the mammalian inner ear: implications for deafness.Mamm Genome. 2009 Sep-Oct;20(9-10):581-603. doi: 10.1007/s00335-009-9230-5. Epub 2009 Oct 30. Mamm Genome. 2009. PMID: 19876605 Review.
-
The pleiotropic transcriptional regulator COUP-TFI plays multiple roles in neural development and disease.Brain Res. 2019 Feb 15;1705:75-94. doi: 10.1016/j.brainres.2018.04.024. Epub 2018 Apr 27. Brain Res. 2019. PMID: 29709504 Review.
Cited by
-
Nr2f1 enhancers have distinct functions in controlling Nr2f1 expression during cortical development.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2402368121. doi: 10.1073/pnas.2402368121. Epub 2024 Sep 23. Proc Natl Acad Sci U S A. 2024. PMID: 39312666 Free PMC article.
-
Mapping oto-pharyngeal development in a human inner ear organoid model.Development. 2023 Oct 1;150(19):dev201871. doi: 10.1242/dev.201871. Epub 2023 Oct 5. Development. 2023. PMID: 37796037 Free PMC article.
-
Characterization of rare spindle and root cell transcriptional profiles in the stria vascularis of the adult mouse cochlea.Sci Rep. 2020 Oct 22;10(1):18100. doi: 10.1038/s41598-020-75238-8. Sci Rep. 2020. PMID: 33093630 Free PMC article.
-
The DNA methylation-regulated MCTP1 activates the drug-resistance of esophageal cancer cells.Aging (Albany NY). 2021 Feb 11;13(3):3342-3352. doi: 10.18632/aging.104173. Epub 2021 Feb 11. Aging (Albany NY). 2021. PMID: 33571139 Free PMC article.
References
-
- Armentano M, Chou SJ, Tomassy GS, Leingartner A, O’Leary DD, Studer M, 2007. COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nature neuroscience 10, 1277–1286. - PubMed
-
- Armentano M, Filosa A, Andolfr G, Studer M, 2006. COUP-TFI is required for the formation of commissural projections in the forebrain by regulating axonal growth. Development 133, 4151–4162. - PubMed
-
- Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G, 2006. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Developmental biology 291, 382–397. - PubMed
-
- Bertacchi M, Parisot J, Studer M, 2018. The pleiotropic transcriptional regulator COUP-TFI plays multiple roles in neural development and disease Brain research. - PubMed
-
- Boettger T, Hubner CA, Maier H, Rust MB, Beck FX, Jentsch TJ, 2002. Deafness and renal tubular acidosis in mice lacking the K-Cl co-transporter Kcc4. Nature 416, 874–878. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

