Two distinct conformations of factor H regulate discrete complement-binding functions in the fluid phase and at cell surfaces
- PMID: 30217822
- PMCID: PMC6222095
- DOI: 10.1074/jbc.RA118.004767
Two distinct conformations of factor H regulate discrete complement-binding functions in the fluid phase and at cell surfaces
Abstract
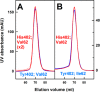
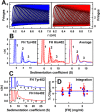
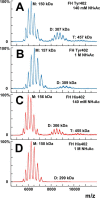
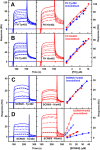
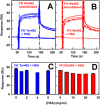
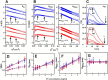
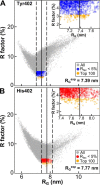
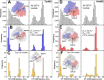
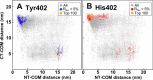
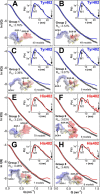
Factor H (FH) is the major regulator of C3b in the alternative pathway of the complement system in immunity. FH comprises 20 short complement regulator (SCR) domains, including eight glycans, and its Y402H polymorphism predisposes those who carry it to age-related macular degeneration. To better understand FH complement binding and self-association, we have studied the solution structures of both the His-402 and Tyr-402 FH allotypes. Analytical ultracentrifugation revealed that up to 12% of both FH allotypes self-associate, and this was confirmed by small-angle X-ray scattering (SAXS), MS, and surface plasmon resonance analyses. SAXS showed that monomeric FH has a radius of gyration (Rg ) of 7.2-7.8 nm and a length of 25 nm. Starting from known structures for the SCR domains and glycans, the SAXS data were fitted using Monte Carlo methods to determine atomistic structures of monomeric FH. The analysis of 29,715 physically realistic but randomized FH conformations resulted in 100 similar best-fit FH structures for each allotype. Two distinct molecular structures resulted that showed either an extended N-terminal domain arrangement with a folded-back C terminus or an extended C terminus and a folded-back N terminus. These two structures are the most accurate to date for glycosylated full-length FH. To clarify FH functional roles in host protection, crystal structures for the FH complexes with C3b and C3dg revealed that the extended N-terminal conformation accounted for C3b fluid-phase regulation, the extended C-terminal conformation accounted for C3d binding, and both conformations accounted for bivalent FH binding to glycosaminoglycans on the target cell surface.
Keywords: C3b activity; Monte Carlo modeling; X-ray scattering; analytical ultracentrifugation; complement; complement regulation; immune system; innate immunity; molecular modeling; solution structure; surface plasmon resonance (SPR).
© 2018 Osborne et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Law S. K., and Reid K. B. (1995) in Complement (Male D., ed) 2nd Ed., IRL Press at Oxford University Press, Oxford, UK
-
- Pangburn M. K., Schreiber R. D., and Müller-Eberhard H. J. (1977) Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein β1H for cleavage of C3b and C4b in solution. J. Exp. Med. 146, 257–270 10.1084/jem.146.1.257 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

