Physical basis for long-distance communication along meiotic chromosomes
- PMID: 30217891
- PMCID: PMC6176642
- DOI: 10.1073/pnas.1801920115
Physical basis for long-distance communication along meiotic chromosomes
Abstract
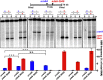
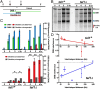
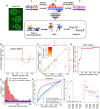
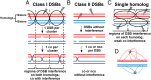
Viable gamete formation requires segregation of homologous chromosomes connected, in most species, by cross-overs. DNA double-strand break (DSB) formation and the resulting cross-overs are regulated at multiple levels to prevent overabundance along chromosomes. Meiotic cells coordinate these events between distant sites, but the physical basis of long-distance chromosomal communication has been unknown. We show that DSB hotspots up to ∼200 kb (∼35 cM) apart form clusters via hotspot-binding proteins Rec25 and Rec27 in fission yeast. Clustering coincides with hotspot competition and interference over similar distances. Without Tel1 (an ATM tumor-suppressor homolog), DSB and crossover interference become negative, reflecting coordinated action along a chromosome. These results indicate that DSB hotspots within a limited chromosomal region and bound by their protein determinants form a clustered structure that, via Tel1, allows only one DSB per region. Such a "roulette" process within clusters explains the observed pattern of crossover interference in fission yeast. Key structural and regulatory components of clusters are phylogenetically conserved, suggesting conservation of this vital regulation. Based on these observations, we propose a model and discuss variations in which clustering and competition between DSB sites leads to DSB interference and in turn produces crossover interference.
Keywords: DNA break interference; DSB hotspot clustering; S. pombe; crossover interference; meiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. - PubMed
-
- Sturtevant AH. The behavior of the chromosomes as studied through linkage. Z Indukt Abstamm Vererbungsl. 1915;13:234–287.
-
- Loidl J. S. pombe linear elements: The modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

