Functional diversification of the NleG effector family in enterohemorrhagic Escherichia coli
- PMID: 30217892
- PMCID: PMC6176568
- DOI: 10.1073/pnas.1718350115
Functional diversification of the NleG effector family in enterohemorrhagic Escherichia coli
Abstract
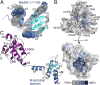
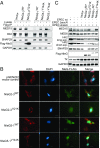
The pathogenic strategy of Escherichia coli and many other gram-negative pathogens relies on the translocation of a specific set of proteins, called effectors, into the eukaryotic host cell during infection. These effectors act in concert to modulate host cell processes in favor of the invading pathogen. Injected by the type III secretion system (T3SS), the effector arsenal of enterohemorrhagic E. coli (EHEC) O157:H7 features at least eight individual NleG effectors, which are also found across diverse attaching and effacing pathogens. NleG effectors share a conserved C-terminal U-box E3 ubiquitin ligase domain that engages with host ubiquitination machinery. However, their specific functions and ubiquitination targets have remained uncharacterized. Here, we identify host proteins targeted for ubiquitination-mediated degradation by two EHEC NleG family members, NleG5-1 and NleG2-3. NleG5-1 localizes to the host cell nucleus and targets the MED15 subunit of the Mediator complex, while NleG2-3 resides in the host cytosol and triggers degradation of Hexokinase-2 and SNAP29. Our structural studies of NleG5-1 reveal a distinct N-terminal α/β domain that is responsible for interacting with host protein targets. The core of this domain is conserved across the NleG family, suggesting this domain is present in functionally distinct NleG effectors, which evolved diversified surface residues to interact with specific host proteins. This is a demonstration of the functional diversification and the range of host proteins targeted by the most expanded effector family in the pathogenic arsenal of E. coli.
Keywords: Escherichia coli; effectors; pathogenesis; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. - PubMed
-
- Caprioli A, Morabito S, Brugère H, Oswald E. Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. - PubMed
-
- Ashida H, Kim M, Sasakawa C. Exploitation of the host ubiquitin system by human bacterial pathogens. Nat Rev Microbiol. 2014;12:399–413. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

