Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization
- PMID: 30218063
- PMCID: PMC6138674
- DOI: 10.1038/s41467-018-06268-0
Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization
Abstract
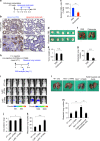
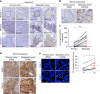
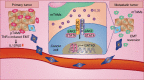
A favorable interplay between cancer cells and the tumor microenvironment (TME) facilitates the outgrowth of metastatic tumors. Because of the distinct initiating processes between primary and metastatic tumors, we investigate the differences in tumor-associated macrophages (TAMs) from primary and metastatic cancers. Here we show that dual expression of M1 and M2 markers is noted in TAMs from primary tumors, whereas predominant expression of M2 markers is shown in metastatic TAMs. At metastatic sites, TAMs secrete interleukin-35 (IL-35) to facilitate metastatic colonization through activation of JAK2-STAT6-GATA3 signaling to reverse epithelial-mesenchymal transition (EMT) in cancer cells. In primary tumors, inflammation-induced EMT upregulates IL12Rβ2, a subunit of the IL-35 receptor, in cancer cells to help them respond to IL-35 during metastasis. Neutralization of IL-35 or knockout of IL-35 in macrophages reduces metastatic colonization. These results indicate the distinct TMEs of primary and metastatic tumors and provide potential targets for intercepting metastasis.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

