Zinc finger nuclease-mediated targeting of multiple transgenes to an endogenous soybean genomic locus via non-homologous end joining
- PMID: 30220095
- PMCID: PMC6419576
- DOI: 10.1111/pbi.13012
Zinc finger nuclease-mediated targeting of multiple transgenes to an endogenous soybean genomic locus via non-homologous end joining
Abstract
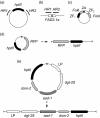
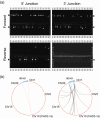
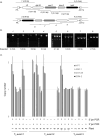
Emerging genome editing technologies hold great promise for the improvement of agricultural crops. Several related genome editing methods currently in development utilize engineered, sequence-specific endonucleases to generate DNA double strand breaks (DSBs) at user-specified genomic loci. These DSBs subsequently result in small insertions/deletions (indels), base substitutions or incorporation of exogenous donor sequences at the target site, depending on the application. Targeted mutagenesis in soybean (Glycine max) via non-homologous end joining (NHEJ)-mediated repair of such DSBs has been previously demonstrated with multiple nucleases, as has homology-directed repair (HDR)-mediated integration of a single transgene into target endogenous soybean loci using CRISPR/Cas9. Here we report targeted integration of multiple transgenes into a single soybean locus using a zinc finger nuclease (ZFN). First, we demonstrate targeted integration of biolistically delivered DNA via either HDR or NHEJ to the FATTY ACID DESATURASE 2-1a (FAD2-1a) locus of embryogenic cells in tissue culture. We then describe ZFN- and NHEJ-mediated, targeted integration of two different multigene donors to the FAD2-1a locus of immature embryos. The largest donor delivered was 16.2 kb, carried four transgenes, and was successfully transmitted to T1 progeny of mature targeted plants obtained via somatic embryogenesis. The insertions in most plants with a targeted, 7.1 kb, NHEJ-integrated donor were perfect or near-perfect, demonstrating that NHEJ is a viable alternative to HDR for gene targeting in soybean. Taken together, these results show that ZFNs can be used to generate fertile transgenic soybean plants with NHEJ-mediated targeted insertions of multigene donors at an endogenous genomic locus.
Keywords: biolistic transformation; gene targeting; genome editing; somatic embryogenesis; soybean; zinc finger nuclease.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

References
-
- Ainley, W.M. , Sastry‐Dent, L. , Welter, M.E. , Murray, M.G. , Zeitler, B. , Amora, R. , Corbin, D.R. et al. (2013) Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134. - PubMed
-
- Ainley, W.M. , Webb, S.R. , Samuel, P. , Guschin, D.Y. , Miller, J.C. and Zhang, L. (2016) FAD2 performance loci and corresponding target site specific binding proteins capable of inducing targeted breaks. US Patent 9, 493, 779.
-
- Barker, R.F. , Idler, K.B. , Thompson, D.V. and Kemp, J.D. (1983) Nucleotide sequence of the T‐DNA region from the Agrobacterium tumefaciens octopine Ti Plasmid pTi15955. Plant Mol. Biol. 2, 335–350. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

