Resequencing core accessions of a pedigree identifies derivation of genomic segments and key agronomic trait loci during cotton improvement
- PMID: 30220108
- PMCID: PMC6419577
- DOI: 10.1111/pbi.13013
Resequencing core accessions of a pedigree identifies derivation of genomic segments and key agronomic trait loci during cotton improvement
Abstract
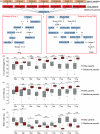
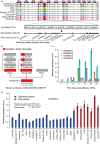
Upland cotton (Gossypium hirsutum) is the world's largest source of natural fibre and dominates the global textile industry. Hybrid cotton varieties exhibit strong heterosis that confers high fibre yields, yet the genome-wide effects of artificial selection that have influenced Upland cotton during its breeding history are poorly understood. Here, we resequenced Upland cotton genomes and constructed a variation map of an intact breeding pedigree comprising seven elite and 19 backbone parents. Compared to wild accessions, the 26 pedigree accessions underwent strong artificial selection during domestication that has resulted in reduced genetic diversity but stronger linkage disequilibrium and higher extents of selective sweeps. In contrast to the backbone parents, the elite parents have acquired significantly improved agronomic traits, with an especially pronounced increase in the lint percentage. Notably, identify by descent (IBD) tracking revealed that the elite parents inherited abundant beneficial trait segments and loci from the backbone parents and our combined analyses led to the identification of a core genomic segment which was inherited in the elite lines from the parents Zhong 7263 and Ejing 1 and that was strongly associated with lint percentage. Additionally, SNP correlation analysis of this core segment showed that a non-synonymous SNP (A-to-G) site in a gene encoding the cell wall-associated receptor-like kinase 3 (GhWAKL3) protein was highly correlated with increased lint percentage. Our results substantially increase the valuable genomics resources available for future genetic and functional genomics studies of cotton and reveal insights that will facilitate yield increases in the molecular breeding of cotton.
Keywords: cotton pedigree; identity by descent (IBD); lint percentage; modern cotton improvement; resequencing.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Bradbury, P.J. , Zhang, Z. , Kroon, D.E. , Casstevens, T.M. , Ramdoss, Y. and Buckler, E.S. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. - PubMed
-
- Brubaker, C.L. , Bourland, F. and Wendel, J.F. (1999) The origin and domestication of cotton. In Cotton: Origin, history, technology, and production ( Wayne Smith, C. , and Tom Cothren, J. , eds), pp. 3–31. New York, NY: John Wiley & Sons.
-
- Campbell, B.T. , Saha, S. , Percy, R. , Frelichowski, J. , Jenkins, J.N. , Park, W. , Mayee, C.D. et al. (2010) Status of the global cotton germplasm resources. Crop Sci. 50, 1161.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

