A Novel Class of ER Membrane Proteins Regulates ER-Associated Endosome Fission
- PMID: 30220460
- PMCID: PMC6195207
- DOI: 10.1016/j.cell.2018.08.030
A Novel Class of ER Membrane Proteins Regulates ER-Associated Endosome Fission
Abstract
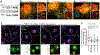
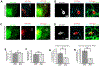
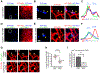
Endoplasmic reticulum (ER) membrane contact sites (MCSs) mark positions where endosomes undergo fission for cargo sorting. To define the role of ER at this unique MCS, we targeted a promiscuous biotin ligase to cargo-sorting domains on endosome buds. This strategy identified the ER membrane protein TMCC1, a member of a conserved protein family. TMCC1 concentrates at the ER-endosome MCSs that are spatially and temporally linked to endosome fission. When TMCC1 is depleted, endosome morphology is normal, buds still form, but ER-associated bud fission and subsequent cargo sorting to the Golgi are impaired. We find that the endosome-localized actin regulator Coronin 1C is required for ER-associated fission of actin-dependent cargo-sorting domains. Coronin 1C is recruited to endosome buds independently of TMCC1, while TMCC1/ER recruitment requires Coronin 1C. This link between TMCC1 and Coronin 1C suggests that the timing of TMCC1-dependent ER recruitment is tightly regulated to occur after cargo has been properly sequestered into the bud.
Keywords: endoplasmic reticulum; endosome fission; endosome recycling; membrane contact site.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures



References
-
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio M-C, et al. (2013). STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J. Cell Sci 126, 5500–5512. - PubMed
-
- Bonifacino JS, and Rojas R (2006). Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol 7, 568–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

