MiR-24-3p regulates cell proliferation and milk protein synthesis of mammary epithelial cells through menin in dairy cows
- PMID: 30221364
- PMCID: PMC6282567
- DOI: 10.1002/jcp.27017
MiR-24-3p regulates cell proliferation and milk protein synthesis of mammary epithelial cells through menin in dairy cows
Abstract
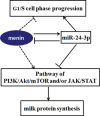
MiR-24-3p, a broadly conserved, small, noncoding RNA, is abundantly expressed in mammary tissue. However, its regulatory role in this tissue remains poorly understood. It was predicted that miR-24-3p targets the 3' untranslated region (3'-UTR) of multiple endocrine neoplasia type 1 (MEN1), an important regulatory factor in mammary tissue. The objective of this study was to investigate the function of miR-24-3p in mammary cells. Using a luciferase assay in mammary epithelial cells (MAC-T), miR-24-3p was confirmed to target the 3'-UTR of MEN1. Furthermore, miR-24-3p negatively regulated the expression of the MEN1 gene and its encoded protein, menin. miR-24-3p enhanced proliferation of MAC-T by promoting G1/S phase progression. MiR-24-3p also regulated the expression of key factors involved in phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin and Janus kinase/signal transducer and activators of transcription signaling pathways, therefore controlling milk protein synthesis in epithelial cells. Thus, miR-24-3p appears to act on MAC-T by targeting MEN1. The expression of miR-24-3p was controlled by MEN1/menin, indicating a negative feedback loop between miR-24-3p and MEN1/menin. The negatively inhibited expression pattern of miR-24-3p and MEN1 was active in mammary tissues at different lactation stages. The feedback mechanism is a new concept to further understand the lactation cycle of mammary glands and can possibly to be manipulated to improve milk yield and quality.
Keywords: cell proliferation; mammary epithelial cells (MAC-T); miR-24-3p; milk protein synthesis; multiple endocrine neoplasia type 1(MEN1)/menin.
© 2018 The Authors. Journal of Cellular Physiology Published by Wiley Periodicals, Inc.
Figures

Similar articles
-
Role of miR-24 in Multiple Endocrine Neoplasia Type 1: A Potential Target for Molecular Therapy.Int J Mol Sci. 2021 Jul 8;22(14):7352. doi: 10.3390/ijms22147352. Int J Mol Sci. 2021. PMID: 34298972 Free PMC article. Review.
-
MEN1/Menin regulates milk protein synthesis through mTOR signaling in mammary epithelial cells.Sci Rep. 2017 Jul 14;7(1):5479. doi: 10.1038/s41598-017-06054-w. Sci Rep. 2017. PMID: 28710500 Free PMC article.
-
Menin Modulates Mammary Epithelial Cell Numbers in Bovine Mammary Glands Through Cyclin D1.J Mammary Gland Biol Neoplasia. 2017 Dec;22(4):221-233. doi: 10.1007/s10911-017-9385-8. Epub 2017 Nov 29. J Mammary Gland Biol Neoplasia. 2017. PMID: 29188494 Free PMC article.
-
MicroRNA-221 regulates proliferation of bovine mammary gland epithelial cells by targeting the STAT5a and IRS1 genes.J Dairy Sci. 2019 Jan;102(1):426-435. doi: 10.3168/jds.2018-15108. Epub 2018 Oct 23. J Dairy Sci. 2019. PMID: 30366615
-
Menin signaling and therapeutic targeting in breast cancer.Curr Probl Cancer. 2024 Aug;51:101118. doi: 10.1016/j.currproblcancer.2024.101118. Epub 2024 Jul 4. Curr Probl Cancer. 2024. PMID: 38968834 Review.
Cited by
-
MicroRNAs in Ruminants and Their Potential Role in Nutrition and Physiology.Vet Sci. 2023 Jan 14;10(1):57. doi: 10.3390/vetsci10010057. Vet Sci. 2023. PMID: 36669058 Free PMC article. Review.
-
Epigenetics: New Insights into Mammary Gland Biology.Genes (Basel). 2021 Feb 5;12(2):231. doi: 10.3390/genes12020231. Genes (Basel). 2021. PMID: 33562534 Free PMC article. Review.
-
Role of miR-24 in Multiple Endocrine Neoplasia Type 1: A Potential Target for Molecular Therapy.Int J Mol Sci. 2021 Jul 8;22(14):7352. doi: 10.3390/ijms22147352. Int J Mol Sci. 2021. PMID: 34298972 Free PMC article. Review.
-
Proteome analysis identified proteins associated with mitochondrial function and inflammation activation crucially regulating the pathogenesis of fatty liver disease.BMC Genomics. 2021 Sep 4;22(1):640. doi: 10.1186/s12864-021-07950-2. BMC Genomics. 2021. PMID: 34481473 Free PMC article.
-
RNA-Seq Reveals the Role of miR-29c in Regulating Inflammation and Oxidative Stress of Bovine Mammary Epithelial Cells.Front Vet Sci. 2022 Apr 1;9:865415. doi: 10.3389/fvets.2022.865415. eCollection 2022. Front Vet Sci. 2022. PMID: 35433915 Free PMC article.
References
-
- Appuhamy, J. A. , Nayananjalie, W. A. , England, E. M. , Gerrard, D. E. , Akers, R. M. , & Hanigan, M. D. (2014). Effects of AMP‐activated protein kinase (AMPK) signaling and essential amino acids on mammalian target of rapamycin (mTOR) signaling and protein synthesis rates in mammary cells. Journal of Dairy Science, 97, 419–429. - PubMed
-
- Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. - PubMed
-
- Bertolino, P. , Tong, W. M. , Galendo, D. , Wang, Z. Q. , & Zhang, C. X. (2003). Heterozygous MEN1 mutant mice develop a range of endocrine tumors mimicking multiple endocrine neoplasia type 1. Molecular Endocrinology, 17(9), 1880–1892. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

