Metformin blocks MYC protein synthesis in colorectal cancer via mTOR-4EBP-eIF4E and MNK1-eIF4G-eIF4E signaling
- PMID: 30221473
- PMCID: PMC6210051
- DOI: 10.1002/1878-0261.12384
Metformin blocks MYC protein synthesis in colorectal cancer via mTOR-4EBP-eIF4E and MNK1-eIF4G-eIF4E signaling
Abstract
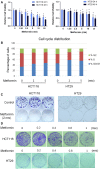
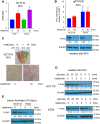
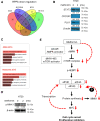
The antidiabetic drug metformin has been associated with reduced colorectal cancer (CRC) risk and improved prognosis of CRC patients. However, the detailed mechanisms underlying such beneficial effects remain unknown. In this study, we aimed to evaluate metformin activity in CRC models and unveil the underlying molecular mechanisms. We showed that metformin inhibits CRC cell proliferation by arresting cells in the G1 phase of the cell cycle and dramatically reduces colony formation of CRC cells. We discovered that metformin causes a robust reduction of MYC protein level. Through the use of luciferase assay and coincubation with either protein synthesis or proteasome inhibitors, we demonstrated that regulation of MYC by metformin is independent of the proteasome and 3' UTR-mediated regulation, but depends on protein synthesis. Data from polysome profiling and ribopuromycylation assays showed that metformin induced widespread inhibition of protein synthesis. Repression of protein synthesis by metformin preferentially affects cell cycle-associated proteins, by altering signaling through the mTOR-4EBP-eIF4E and MNK1-eIF4G-eIF4E axes. The inhibition of MYC protein synthesis may underlie metformin's beneficial effects on CRC risk and prognosis.
Keywords: MYC; mTOR; cell cycle; colorectal cancer; metformin; protein synthesis.
© 2018 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures



References
-
- Blandino G, Valerio M, Cioce M, Mori F, Casadei L, Pulito C, Sacconi A, Biagioni F, Cortese G, Galanti S et al (2012) Metformin elicits anticancer effects through the sequential modulation of DICER and c‐MYC. Nat Commun 3, 865. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

