Using Illumina Infinium HumanMethylation 450K BeadChip to explore genome‑wide DNA methylation profiles in a human hepatocellular carcinoma cell line
- PMID: 30221710
- PMCID: PMC6172394
- DOI: 10.3892/mmr.2018.9441
Using Illumina Infinium HumanMethylation 450K BeadChip to explore genome‑wide DNA methylation profiles in a human hepatocellular carcinoma cell line
Abstract
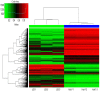
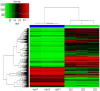
Aberrant DNA methylation is the most common type of epigenetic alteration and is associated with many types of cancer. Although previous studies have provided a few novel DNA methylation markers in hepatocellular carcinoma (HCC), specific DNA methylation patterns and comparisons of the aberrant alterations in methylation between HCC and normal liver cell lines have not yet been reported. Therefore, in the present study the Illumina Infinium HumanMethylation 450K BeadChip was employed to identify the genome‑wide aberrant DNA methylation profiles of Huh7 and L02 cells. Following Bonferroni adjustment, 102,254 differentially methylated CpG sites (covering 26,511 genes) were detected between Huh7 and L02 cells. Of those CpG sites, 62,702 (61.3%) sites were hypermethylated (covering 12,665 genes) and 39,552 (38.7%) sites were hypomethylated (covering 13,846 genes). The results of the present study indicated that 40.3% of the CpG sites were in CpG island regions, 20.7% were in CpG shores and 8.8% were in shelf regions. A total of 57.3% hypermethylated CpG sites and 39.4% of the hypomethylated CpG sites had a |β‑Difference| ≥50%. Within the significant differentially methylated CpG sites, 490 genes were located within 598 differentially methylated regions. Gene Ontology enrichment analysis revealed that 2,107 differentially methylated genes were associated with 'biological process', 13,351 differentially methylated genes were associated with 'molecular function', and 18,041 differentially methylated genes were associated with 'cellular component'. Kyoto Encyclopedia of Genes and Genomes pathway‑based analysis revealed 43 signaling pathways that were associated with 5,195 differentially methylated genes. These results demonstrated that aberrant DNA methylation may be a key and common event underlying the tumorigenesis of Huh7 cells. The present study also identified many subsets of hypo‑ or hyper‑methylated CpG sites, genes and signaling pathways, which have an importance in the occurrence and development of HCC.
Figures


Similar articles
-
Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips.Epigenetics. 2013 Jan;8(1):34-43. doi: 10.4161/epi.23062. Epub 2012 Dec 3. Epigenetics. 2013. PMID: 23208076 Free PMC article.
-
Epigenomic profiling of DNA methylation for hepatocellular carcinoma diagnosis and prognosis prediction.J Gastroenterol Hepatol. 2019 Oct;34(10):1869-1877. doi: 10.1111/jgh.14694. Epub 2019 Jun 20. J Gastroenterol Hepatol. 2019. PMID: 31038805
-
Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma.PLoS One. 2013;8(2):e55761. doi: 10.1371/journal.pone.0055761. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437062 Free PMC article.
-
Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma.World J Gastroenterol. 2015 May 28;21(20):6317-28. doi: 10.3748/wjg.v21.i20.6317. World J Gastroenterol. 2015. PMID: 26034368 Free PMC article. Review.
-
Comparison of genome-wide analysis techniques to DNA methylation analysis in human cancer.J Cell Physiol. 2018 May;233(5):3968-3981. doi: 10.1002/jcp.26176. Epub 2017 Oct 4. J Cell Physiol. 2018. PMID: 28888056 Review.
Cited by
-
Epigenetic modifications and the development of kidney graft fibrosis.Curr Opin Organ Transplant. 2021 Feb 1;26(1):1-9. doi: 10.1097/MOT.0000000000000839. Curr Opin Organ Transplant. 2021. PMID: 33315766 Free PMC article. Review.
-
Insights into the Roles of Epigenetic Modifications in Ferroptosis.Biology (Basel). 2024 Feb 15;13(2):122. doi: 10.3390/biology13020122. Biology (Basel). 2024. PMID: 38392340 Free PMC article. Review.
-
Genome-Wide Scan for Copy Number Variations in Chinese Merino Sheep Based on Ovine High-Density 600K SNP Arrays.Animals (Basel). 2024 Oct 8;14(19):2897. doi: 10.3390/ani14192897. Animals (Basel). 2024. PMID: 39409846 Free PMC article.
-
The Relevance of SOCS1 Methylation and Epigenetic Therapy in Diverse Cell Populations of Hepatocellular Carcinoma.Diagnostics (Basel). 2021 Oct 2;11(10):1825. doi: 10.3390/diagnostics11101825. Diagnostics (Basel). 2021. PMID: 34679523 Free PMC article.
-
Epigenetic silencing of MEIS2 in prostate cancer recurrence.Clin Epigenetics. 2019 Oct 22;11(1):147. doi: 10.1186/s13148-019-0742-x. Clin Epigenetics. 2019. PMID: 31640805 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

