The Spastic Paraplegia-Associated Phospholipase DDHD1 Is a Primary Brain Phosphatidylinositol Lipase
- PMID: 30221923
- PMCID: PMC6237197
- DOI: 10.1021/acs.biochem.8b00810
The Spastic Paraplegia-Associated Phospholipase DDHD1 Is a Primary Brain Phosphatidylinositol Lipase
Abstract
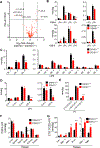
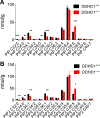
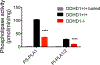
Deleterious mutations in the serine hydrolase DDHD domain containing 1 (DDHD1) cause the SPG28 subtype of the neurological disease hereditary spastic paraplegia (HSP), which is characterized by axonal neuropathy and gait impairments. DDHD1 has been shown to display PLA1-type phospholipase activity with a preference for phosphatidic acid. However, the endogenous lipid pathways regulated by DDHD1 in vivo remain poorly understood. Here we use a combination of untargeted and targeted metabolomics to compare the lipid content of brain tissue from DDHD1+/+ and DDHD1-/- mice, revealing that DDHD1 inactivation causes a substantial decrease in the level of polyunsaturated lysophosphatidylinositol (LPI) lipids and a corresponding increase in the level of phosphatidylinositol (PI) lipids. Levels of other phospholipids were mostly unchanged, with the exception of decreases in the levels of select polyunsaturated lysophosphatidylserine (LPS) and lysophosphatidylcholine lipids and a striking remodeling of PI phosphates (e.g., PIP and PIP2) in DDHD1-/- brain tissue. Biochemical assays confirmed that DDHD1 hydrolyzes PI/PS to LPI/LPS with sn-1 selectivity and accounts for a substantial fraction of the PI/PS lipase activity in mouse brain tissue. These data indicate that DDHD1 is a principal regulator of bioactive LPI and other lysophospholipids, as well as PI phosphates, in the mammalian nervous system, pointing to a potential role for these lipid pathways in HSP.
Figures

References
-
- Bisogno T; Howell F; Williams G; Minassi A; Cascio MG; Ligresti A; Matias I; Schiano-Moriello A; Paul P; Williams E-J; Gangadharan U; Hobbs C; Di Marzo V; Doherty P Cloning of the First Sn1-DAG Lipases Points to the Spatial and Temporal Regulation of Endocannabinoid Signaling in the Brain. J. Cell Biol 2003, 163 (3), 463–468. - PMC - PubMed
-
- Yamashita A; Oka S; Tanikawa T; Hayashi Y; Nemoto-Sasaki Y; Sugiura T The Actions and Metabolism of Lysophosphatidylinositol, an Endogenous Agonist for GPR55. Prostaglandins Other Lipid Mediat. 2013, 107, 103–116. - PubMed
-
- Higgs HN; Han MH; Johnson GE; Glomset JA Cloning of a Phosphatidic Acid-Preferring Phospholipase A1 From Bovine Testis. J. Biol. Chem 1998, 273 (10), 5468–5477. - PubMed
-
- Higgs HN; Glomset JA Purification and Properties of a Phosphatidic Acid-Preferring Phospholipase A1 From Bovine Testis. Examination of the Molecular Basis of Its Activation. J. Biol. Chem 1996, 271 (18), 10874–10883. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

