Signaling pathways as linear transmitters
- PMID: 30222104
- PMCID: PMC6202053
- DOI: 10.7554/eLife.33617
Signaling pathways as linear transmitters
Abstract
One challenge in biology is to make sense of the complexity of biological networks. A good system to approach this is signaling pathways, whose well-characterized molecular details allow us to relate the internal processes of each pathway to their input-output behavior. In this study, we analyzed mathematical models of three metazoan signaling pathways: the canonical Wnt, MAPK/ERK, and Tgfβ pathways. We find an unexpected convergence: the three pathways behave in some physiological contexts as linear signal transmitters. Testing the results experimentally, we present direct measurements of linear input-output behavior in the Wnt and ERK pathways. Analytics from each model further reveal that linearity arises through different means in each pathway, which we tested experimentally in the Wnt and ERK pathways. Linearity is a desired property in engineering where it facilitates fidelity and superposition in signal transmission. Our findings illustrate how cells tune different complex networks to converge on the same behavior.
Keywords: cell biology; computational biology; human cell lines; linearity; signal transmission; signaling pathways; systems biology.
© 2018, Nunns et al.
Conflict of interest statement
HN, LG No competing interests declared
Figures
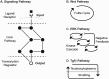

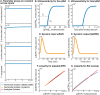
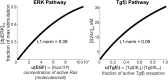




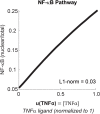

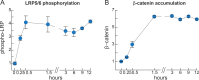
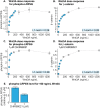


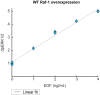





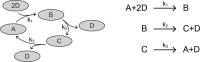
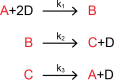
Comment in
-
Transferring information without distortion.Elife. 2018 Oct 25;7:e41894. doi: 10.7554/eLife.41894. Elife. 2018. PMID: 30358530 Free PMC article.
References
-
- Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DB, Rand DA, Sée V, White MR. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

