Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells
- PMID: 30222169
- PMCID: PMC6546596
- DOI: 10.1038/nbt.4231
Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells
Abstract
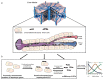
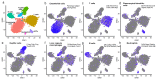
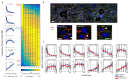
Spatially resolved single-cell RNA sequencing (scRNAseq) is a powerful approach for inferring connections between a cell's identity and its position in a tissue. We recently combined scRNAseq with spatially mapped landmark genes to infer the expression zonation of hepatocytes. However, determining zonation of small cells with low mRNA content, or without highly expressed landmark genes, remains challenging. Here we used paired-cell sequencing, in which mRNA from pairs of attached mouse cells were sequenced and gene expression from one cell type was used to infer the pairs' tissue coordinates. We applied this method to pairs of hepatocytes and liver endothelial cells (LECs). Using the spatial information from hepatocytes, we reconstructed LEC zonation and extracted a landmark gene panel that we used to spatially map LEC scRNAseq data. Our approach revealed the expression of both Wnt ligands and the Dkk3 Wnt antagonist in distinct pericentral LEC sub-populations. This approach can be used to reconstruct spatial expression maps of non-parenchymal cells in other tissues.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Regev A, et al. The Human Cell Atlas. bioRxiv. 2017 doi: 10.1101/121202. 121202. - DOI
-
- Transcriptomic characterization of 20 organs and tissues from mouse at single cell resolution creates a Tabula Muris. [Accessed: 27th December 2017];bioRxiv. Available at: https://www.biorxiv.org/content/early/2017/12/20/237446.
-
- Han X, et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. - PubMed
-
- Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. - PubMed
-
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The Technology and Biology of Single-Cell RNA Sequencing. Molecular cell. 2015;58:610–620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

