The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex
- PMID: 30222246
- PMCID: PMC6398391
- DOI: 10.1111/febs.14660
The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex
Abstract
c-MYC and the SWI/SNF chromatin remodeling complex act as master regulators of transcription, and play a key role in human cancer. Although they are known to interact, the molecular details of their interaction are lacking. We have determined the structure of the RPT1 region of the INI1/hSNF5/BAF47/SMARCB1 subunit of the SWI/SNF complex that acts as a c-MYC-binding domain, and have localized the interaction regions on both INI1 and on the c-MYC:MAX heterodimer. c-MYC interacts with a highly conserved groove on INI1, while INI1 binds to the c-MYC helix-loop-helix region. The binding site overlaps with the c-MYC DNA-binding region, and we show that binding of INI1 and E-box DNA to c-MYC:MAX are mutually exclusive.
Keywords: MYC; BAF/PBAF complexes; INI1/hSNF5/BAF47/SMARCB1 subunit; protein-protein interactions; transcription factors.
© 2018 Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



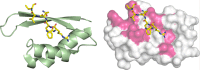






Similar articles
-
MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation.Cell Cycle. 2016 Jul 2;15(13):1693-705. doi: 10.1080/15384101.2016.1146836. Epub 2016 Jun 7. Cell Cycle. 2016. PMID: 27267444 Free PMC article.
-
c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function.Nat Genet. 1999 May;22(1):102-5. doi: 10.1038/8811. Nat Genet. 1999. PMID: 10319872
-
Computational modeling of Repeat1 region of INI1/hSNF5: An evolutionary link with ubiquitin.Protein Sci. 2016 Sep;25(9):1593-604. doi: 10.1002/pro.2961. Epub 2016 Jun 27. Protein Sci. 2016. PMID: 27261671 Free PMC article.
-
The role of INI1/hSNF5 in gene regulation and cancer.Biochem Cell Biol. 2009 Feb;87(1):163-77. doi: 10.1139/O08-113. Biochem Cell Biol. 2009. PMID: 19234532 Review.
-
Integrase interactor 1 in health and disease.Curr Protein Pept Sci. 2015;16(6):478-90. doi: 10.2174/1389203716666150316115156. Curr Protein Pept Sci. 2015. PMID: 25772157 Review.
Cited by
-
Exploring the dynamics and interactions of the N-myc transactivation domain through solution nuclear magnetic resonance spectroscopy.Biochem J. 2024 Nov 6;481(21):1535-1556. doi: 10.1042/BCJ20240248. Biochem J. 2024. PMID: 39370942 Free PMC article.
-
SWI/SNF complexes in hematological malignancies: biological implications and therapeutic opportunities.Mol Cancer. 2023 Feb 21;22(1):39. doi: 10.1186/s12943-023-01736-8. Mol Cancer. 2023. PMID: 36810086 Free PMC article. Review.
-
Structural and Biophysical Insights into the Function of the Intrinsically Disordered Myc Oncoprotein.Cells. 2020 Apr 22;9(4):1038. doi: 10.3390/cells9041038. Cells. 2020. PMID: 32331235 Free PMC article. Review.
-
Structure of the BRK domain of the SWI/SNF chromatin remodeling complex subunit BRG1 reveals a potential role in protein-protein interactions.Protein Sci. 2020 Apr;29(4):1047-1053. doi: 10.1002/pro.3820. Epub 2020 Jan 13. Protein Sci. 2020. PMID: 31909846 Free PMC article.
-
Epigenetic Regulation in Primary CNS Tumors: An Opportunity to Bridge Old and New WHO Classifications.Cancers (Basel). 2023 Apr 27;15(9):2511. doi: 10.3390/cancers15092511. Cancers (Basel). 2023. PMID: 37173979 Free PMC article. Review.
References
-
- Zinzalla G (2016) Targeting MYC: is it getting any easier? Future Med Chem 8, 1899–1902. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

