ENDOR Characterization of (N2)FeII(μ-H)2FeI(N2)-: A Spectroscopic Model for N2 Binding by the Di-μ-hydrido Nitrogenase Janus Intermediate
- PMID: 30222330
- PMCID: PMC6649688
- DOI: 10.1021/acs.inorgchem.8b02021
ENDOR Characterization of (N2)FeII(μ-H)2FeI(N2)-: A Spectroscopic Model for N2 Binding by the Di-μ-hydrido Nitrogenase Janus Intermediate
Abstract
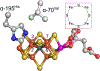
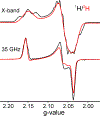
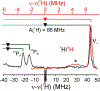
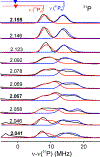
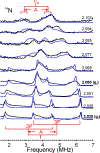
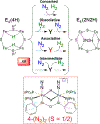
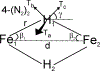
The biomimetic diiron complex 4-(N2)2, featuring two terminally bound Fe-N2 centers bridged by two hydrides, serves as a model for two possible states along the pathway by which the enzyme nitrogenase reduces N2. One is the Janus intermediate E4(4H), which has accumulated 4[e-/H+], stored as two [Fe-H-Fe] bridging hydrides, and is activated to bind and reduce N2 through reductive elimination (RE) of the hydride ligands as H2. The second is a possible RE intermediate. 1H and 14N 35 GHz ENDOR measurements confirm that the formally Fe(II)/Fe(I) 4-(N2)2 complex exhibits a fully delocalized, Robin-Day type-III mixed valency. The two bridging hydrides exhibit a fully rhombic dipolar tensor form, T ≈ [- t, + t, 0]. The rhombic form is reproduced by a simple point-dipole model for dipolar interactions between a bridging hydride and its "anchor" Fe ions, confirming validity of this model and demonstrating that observation of a rhombic form is a convenient diagnostic signature for the identification of such core structures in biological centers such as nitrogenase. Furthermore, interpretation of the 1H measurements with the anchor model maps the g tensor onto the molecular frame, an important function of these equations for application to nitrogenase. Analysis of the hyperfine and quadrupole coupling to the bound 14N of N2 provides a reference for nitrogen-bound nitrogenase intermediates and is of chemical significance, as it gives a quantitative estimate of the amount of charge transferred between Fe and coordinated N, a key element in N2 activation for reduction.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
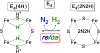



Similar articles
-
High-Resolution ENDOR Spectroscopy Combined with Quantum Chemical Calculations Reveals the Structure of Nitrogenase Janus Intermediate E4(4H).J Am Chem Soc. 2019 Jul 31;141(30):11984-11996. doi: 10.1021/jacs.9b04474. Epub 2019 Jul 16. J Am Chem Soc. 2019. PMID: 31310109 Free PMC article.
-
Modeling the signatures of hydrides in metalloenzymes: ENDOR analysis of a Di-iron Fe(μ-NH)(μ-H)Fe core.J Am Chem Soc. 2012 Aug 1;134(30):12637-47. doi: 10.1021/ja303739g. Epub 2012 Jul 23. J Am Chem Soc. 2012. PMID: 22823933 Free PMC article.
-
Reductive Elimination of H2 Activates Nitrogenase to Reduce the N≡N Triple Bond: Characterization of the E4(4H) Janus Intermediate in Wild-Type Enzyme.J Am Chem Soc. 2016 Aug 24;138(33):10674-83. doi: 10.1021/jacs.6b06362. Epub 2016 Aug 16. J Am Chem Soc. 2016. PMID: 27529724 Free PMC article.
-
Insight into the Iron-Molybdenum Cofactor of Nitrogenase from Synthetic Iron Complexes with Sulfur, Carbon, and Hydride Ligands.J Am Chem Soc. 2016 Jun 15;138(23):7200-11. doi: 10.1021/jacs.6b00747. Epub 2016 Jun 3. J Am Chem Soc. 2016. PMID: 27171599 Free PMC article. Review.
-
Breaking the N2 triple bond: insights into the nitrogenase mechanism.Dalton Trans. 2006 May 21;(19):2277-84. doi: 10.1039/b517633f. Epub 2006 Apr 11. Dalton Trans. 2006. PMID: 16688314 Review.
Cited by
-
The Spectroscopy of Nitrogenases.Chem Rev. 2020 Jun 24;120(12):5005-5081. doi: 10.1021/acs.chemrev.9b00650. Epub 2020 Apr 2. Chem Rev. 2020. PMID: 32237739 Free PMC article. Review.
-
Scalar Relativistic All-Electron and Pseudopotential Ab Initio Study of a Minimal Nitrogenase [Fe(SH)4H]- Model Employing Coupled-Cluster and Auxiliary-Field Quantum Monte Carlo Many-Body Methods.J Phys Chem A. 2024 Feb 22;128(7):1358-1374. doi: 10.1021/acs.jpca.3c05808. Epub 2024 Feb 7. J Phys Chem A. 2024. PMID: 38324717 Free PMC article.
-
Structure and dynamics of catalytically competent but labile paramagnetic metal-hydrides: the Ti(iii)-H in homogeneous olefin polymerization.Chem Sci. 2020 Sep 24;11(46):12436-12445. doi: 10.1039/d0sc04967k. Chem Sci. 2020. PMID: 34123229 Free PMC article.
-
Mechanism of Nitrogen Reduction to Ammonia in a Diiron Model of Nitrogenase.Inorg Chem. 2023 Sep 11;62(36):14715-14726. doi: 10.1021/acs.inorgchem.3c02089. Epub 2023 Aug 31. Inorg Chem. 2023. PMID: 37650683 Free PMC article.
-
High-Resolution ENDOR Spectroscopy Combined with Quantum Chemical Calculations Reveals the Structure of Nitrogenase Janus Intermediate E4(4H).J Am Chem Soc. 2019 Jul 31;141(30):11984-11996. doi: 10.1021/jacs.9b04474. Epub 2019 Jul 16. J Am Chem Soc. 2019. PMID: 31310109 Free PMC article.
References
-
- Burgess BK; Lowe DJ Mechanism of Molybdenum Nitrogenase. Chem Rev 1996, 96, 2983–3012. - PubMed
-
- Christiansen J; Seefeldt LC; Dean DR Competitive Substrate and Inhibitor Interactions at the Physiologically Relevant Active Site of Nitrogenase. J. Biol. Chem 2000, 275, 36104–36107. - PubMed
-
- Kim C-H; Newton WE; Dean DR Role of the Mofe Protein α-Subunit Histidine-195 Residue in Femo-Cofactor Binding and Nitrogenase Catalysis. Biochemistry 1995, 34, 2798–2808. - PubMed
-
- Barney BM; Igarashi RY; Dos Santos PC; Dean DR; Seefeldt LC Substrate Interaction at an Iron-Sulfur Face of the Femo-Cofactor During Nitrogenase Catalysis. J. Biol. Chem 2004, 279, 53621–53624. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

